A research group led by Professor Hiroyuki Kurata of the Graduate School of Information Engineering, Kyushu Institute of Technology has succeeded in using an independently developed deep learning model that utilizes only protein amino acid sequence information to achieve the world's highest level of accurate results for the rate of human-virus protein-protein interactions (HV-PPI).
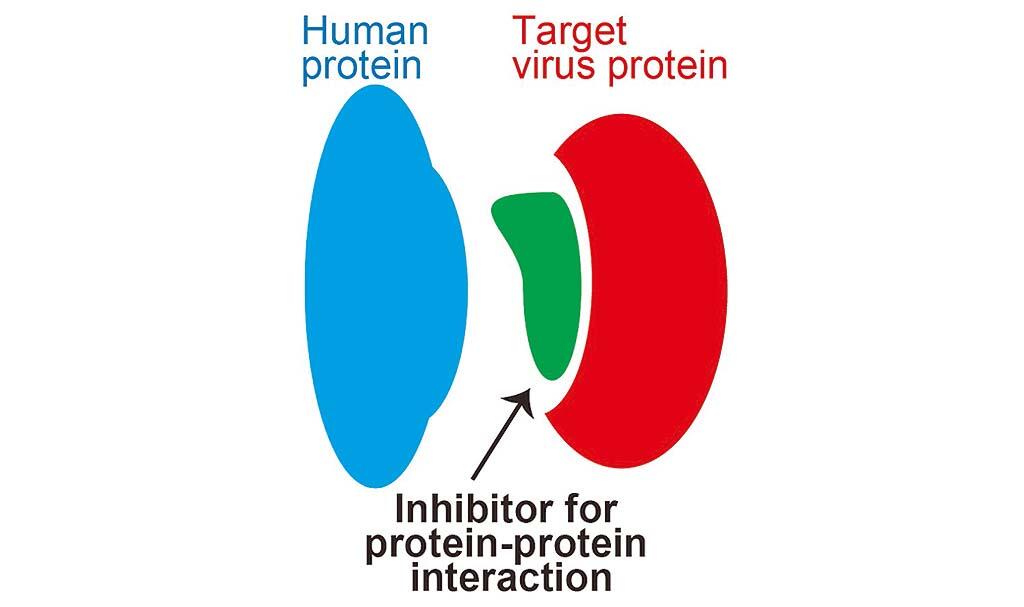
Drug targets for small molecular compounds are often intracellular enzymes, and those for antibody drugs are often cell-surface receptor proteins. However, one looming issue is that we are close to running out of the known drug discovery targets. In order to dramatically increase drug discovery targets, inhibiting intracellular protein-protein interactions has been proposed. Conversely, in vivo experiments to identify these protein-protein interactions have been a very difficult and expensive challenge to overcome.
To tackle this issue, the research group decided to take up the challenge of predicting the intracellular protein-protein interactions from amino acid sequence information using artificial intelligence. The researchers thought that the problem could be overcome through considering the amino acid sequence of a protein as a flow of context and extracting the three-dimensional features of the key and lock using the order pattern of the long-chain amino acid sequence.
To implement this idea, they applied a deep-layer learning model, LSTM, that predicts the futuristic outcomes from time-series data and a distributed representation technique of words in natural language, word2vec. As a consequence, a deep-learning model was developed through combining both LSTM and word2vec, which could accurately capture the stereological features of the key and lock from the amino acid sequence and predict HV-PPI at the highest global level with an accurate rate of 98%. The findings were published online on June 23, 2021 in Briefings in Bioinformatics of the University of Oxford Publishing Agency.
"Using the artificial intelligence technology developed in this study, we aim to identify peptide drugs that can be used to predict the interaction between SARS-CoV-2 and human proteins and inhibit the binding of both proteins," as Professor Kurata said. "Protein interactions are mainly inhibited using medium-sized molecules, such as peptides, rather than conventional small-molecule compounds, but we wish to apply them in developing drugs for other infectious diseases and cancers."
Credit: Kyushu Institute of Technology
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




