Associate Professor Shinichi Arimura and graduate student Issei Nakazato from the Graduate School of Agricultural and Life Sciences, The University of Tokyo as well as a research group at Kyoto University announced their successful editing of the chloroplast genome of the model plant Arabidopsis thaliana. Through editing the nuclear genome of Arabidopsis thaliana using platinum TALEN, they were able to create a plant in which only a specific base of the chloroplast genome was substituted with another base. They also confirmed that these traits were passed on to future generations. Furthermore, they succeeded in obtaining a plant that does not have any remnant marker genes introduced into the nucleus through the editing process, and therefore, is not regarded as a genetically modified organism in later generations. It is an unexpected result that could allow breeding through modifying the chloroplast genome in the future. The results were published in the July 2021 issue of the international journal Nature Plants.
Chloroplasts, the organelles responsible for photosynthesis in plant cells, are thought to originate from cells that coexisted billions of years ago, similar to mitochondria. Many of the chloroplast genes were either lost or aggregated in the nuclear genome. Currently, only 1-2% of these genes exist, and those important for photosynthesis still remain in the chloroplast. As chloroplast DNA is maternally inherited, modifications are expected to be realized and it is expected that the efficiency of photosynthesis along with the yield and biomass of crops can be improved through their editing.
Until now, genetic modification of the chloroplast genome has not been performed in model plants because its applicability is limited to only some plants, such as tobacco. Moreover, even with genome editing technology, modifying all the chloroplast genes was thought to be difficult owing to the extremely high copy number of the chloroplast genome. The research group succeeded in modifying the mitochondrial genome in rice and rapeseed in 2019 and in Arabidopsis thaliana the following year, thereby establishing the editing technology.
In this research, the group applied the method for modifying the mitochondrial genome to the chloroplast. Specifically, a genome-editing protein called "plastid-targeted platinum TALE cytidine deaminase (ptpTALECD)" was designed, in which a chloroplast-targeting signal sequence and an enzyme that converts the DNA base cytosine (C) to thymine (T) are fused to the DNA-binding domain of the artificial restriction enzyme Platinum TALEN. The DNA sequence of this construct was introduced into the nuclear genome of Arabidopsis thaliana.
The objective of using this DNA sequence was to translocate the intracellularly produced ptpTALECD to the chloroplast, which would bind the two TALE domains to the target sequence (16S rRNA) on the chloroplast genome and modify a specific C between the two TALE domains to a T. They succeeded in obtaining a plant in which a specific C in the chloroplast genome, which has more than 1,000 copies per cell, was completely modified to T.
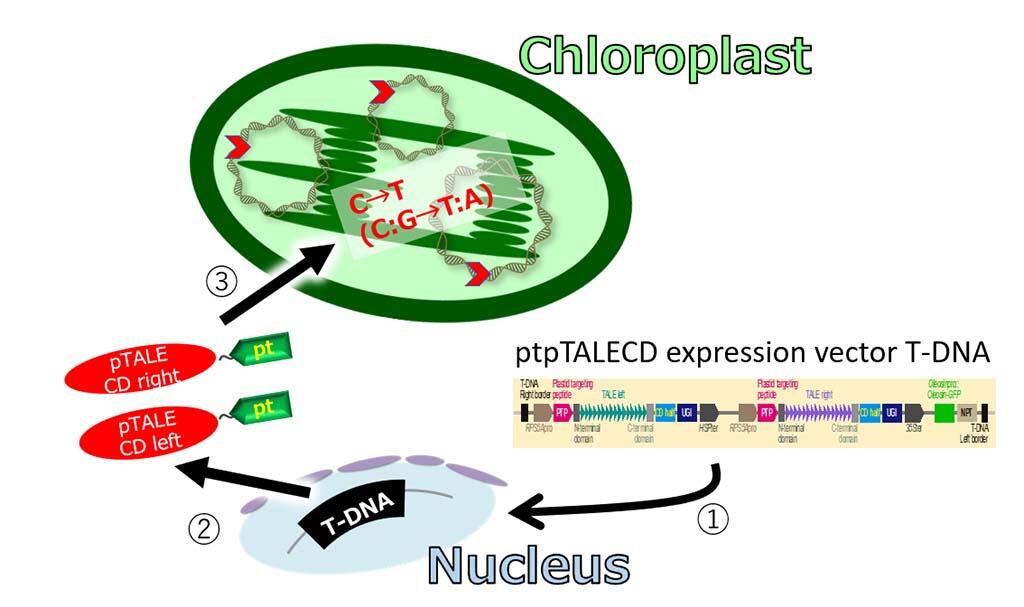
2. The mRNAs were transcribed from the T-DNA and translated in the cytosol into a pair of genome editing enzymes (red; pTALECD left and right) with plastid targeting signals (green tags labeled as "pt" in yellow).
3. The enzyme proteins were transported to the chloroplasts by the sequence information of the "pt" tags and converted all target Cs to Ts of all copies (thousands in each cell!) of the chloroplast genome.
Credit: The University of Tokyo
Through analyzing the plant using a next-generation sequencer, they confirmed that only specific transformations had occurred without any unintended mutations (off-target mutations). To investigate whether the specific mutation was inherited by later generations, plants with the completely substituted specific base were crossed. Antibiotic (spectinomycin) resistance and fluorescent tags added to the transgene were examined in the resulting progeny.
Substitution within the 16S rRNA gene is shown to confer spectinomycin resistance in plants. The mutations possessed by the parents were found to be stably inherited by all individuals. Among these plants, the DNA sequence of ptpTALECD introduced into the parent nuclear genome disappeared, and some individuals did not possess the foreign DNA in their nuclear genome. Since these plants do not fall under the criteria for handling genetically modified plants in Japan, it is expected that this technology can be applied for crop breeding.
Associate Professor Arimura said, "The chloroplast genome accounts for only approximately 1% of all genes, but it contains genes necessary for photosynthesis. We can develop essential technology for practical use in plants, and we believe that we can contribute to CO2 absorption and agricultural production through collaboration with different research groups in Japan and overseas."
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




