The research teams of Associate Professor Akihiko Nakamura, Faculty of Agriculture, Shizuoka University, as well as Associate Professor Nobuyasu Koga and Professor Ryota Iino, Institute for Molecular Science, modified PET2, a polyethylene terephthalate (PET)-degrading enzyme that is widely used worldwide and improved its thermal stability by 6.7 °C and PET degrading activity by 6.8 times. In addition, the structural basis for improving the thermal stability was investigated by X-ray crystallography, and the mechanism for the improved PET degradation activity was elucidated by observing one molecule.
Plastic, which is a synthetic polymer, is inexpensive and easy to process, which has resulted in its mass-production and extensive use in modern society. However, complete recycling of plastics and remediation of environmental pollution caused by plastics remain global concerns toward realizing a sustainable society. In particular, PET is widely used in beverage bottles, clothing, packaging materials, and other applications. Therefore, the research team first used water-soluble bis (2-hydroxyethyl) terephthalate (BHET; an organic compound), which forms a part of the unit structure of PET (polymer solid), as a model substrate, and revealed that the activity of natural PET2 completely diminished by heat treatment at 70 ° C for one hour.
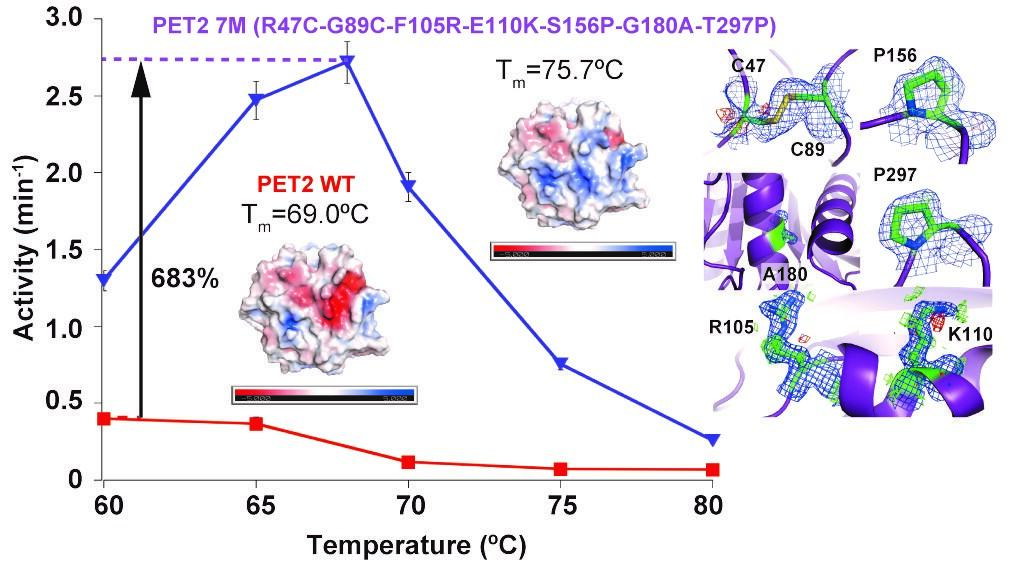
Using the 3D structure obtained through homology modelling, stabilization of the loop structure was achieved by introducing a proline mutation, the helix structure near the catalytic site by introducing an alanine mutation, and the overall structure by introducing a sulfide bond were made. Through this process, PET2 7M with mutations at seven locations was obtained.
The denaturation temperature of this PET2 7M was 75.7 °C, corresponding to an improvement of 6.7 °C over that of natural PET2. Furthermore, the PET degradation activity of PET2 7M at 60 °C was 3.3 times higher than that of natural PET2. In addition, the optimum temperature for PET decomposition changed from 60 °C to 68 °C, an 8 °C increase compared to natural PET2, and the PET degradation activity at each optimum temperature was up to 6.8 times that of the natural PET2.
PET2 7M continued to decompose PET for at least 24 h without denaturation at an optimum temperature of 68 °C. To determine the structural basis for thermal stabilization, the three-dimensional structure of PET2 7M was obtained using X-ray crystallography. Furthermore, it was confirmed that the loop structure did not change and was stabilized by the introduction of the proline mutation, while the helix structure near the catalytic site was stabilized by the introduction of the alanine mutation, and a disulfide bond was indeed formed. The mechanism for the improvement in the degradation activity was also confirmed by observing a single molecule.
Credit: Shizuoka University
PET2 7M was found to bind to the PET surface 3.4 times faster than natural PET2. This difference was similar to the difference in the PET degradation activity (3.3 times) between PET2 7M and natural PET2, when compared at the same temperature (60 °C). The time from binding to PET to dissociation (speed of dissociation) was similar between PET2 7M and natural PET2.
According to Professor Iino: "We have shown that the speed of binding to the PET surface is important for the activation of PET-degrading enzymes. In future investigations, binding to PET will be further accelerated by modifying the enzyme surface and fusion with other proteins. In addition, by improving the heat resistance, the decomposition reaction can be performed above the glass transition temperature of PET. Through these efforts, we hope to further improve the efficiency of PET decomposition and apply it practically."
■Glass transition temperature: The temperature at which a resin such as PET changes from a hard solid (glass) to a soft rubber state.
■Homology modeling: A method of searching for proteins with similar amino acid sequences from proteins whose three-dimensional structures are known and creating a model of the three-dimensional structure based on the search.
■Disulfide bond: A covalent bond formed between the Sulphur atoms in two thiols.
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




