The relationship between brief, dynamic fluctuations in life systems and evolutionary changes over many generations has long been debated as a central theme in life sciences. However, the quantitative verification of evolutionary relationships is difficult and clarifying the theoretical origin of a relationship is usually impossible. Qian-Yuan Tang, Specially appointed researcher (currently at RIKEN), and Professor Kunihiko Kaneko, Graduate School of Arts and Sciences of the University of Tokyo, analyzed protein databases and quantitatively reported a strong correlation between structural fluctuations of proteins and structural changes due to evolution. Furthermore, the origin of these fluctuations was clarified using a theoretical model. These findings were published in Physical Review Letters.
Proteins form the basis of various biological functions, including catalytic activity. Functional proteins have a stable structure, but this structure fluctuates due to the heat around them. These fluctuations can also trigger their functions. On the contrary, evolution leads to mutations in the amino acid sequence and structural deformities.
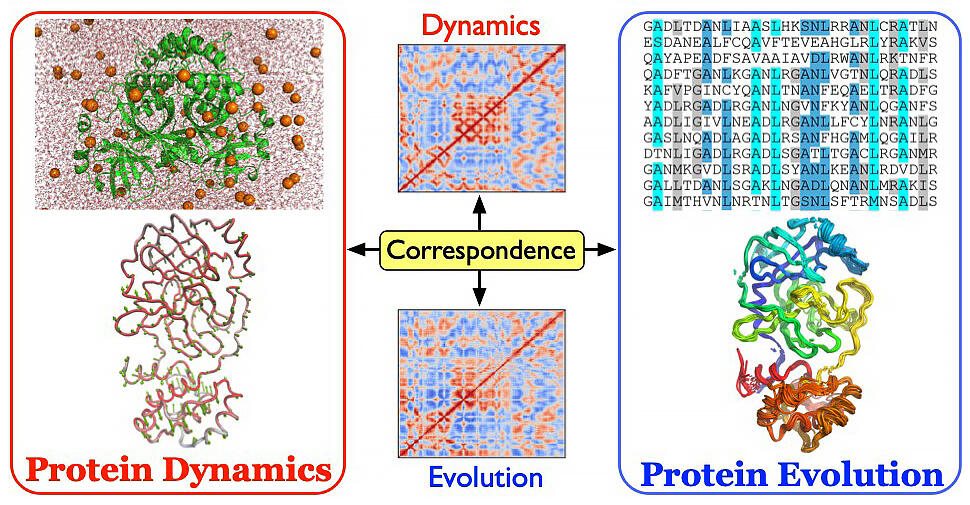
Left: The protein structure fluctuates under thermal noise (top), exhibiting its functional dynamics around the native state (bottom). Right: The mutations in the amino-acid sequences (top) lead to the structural variations of the proteins (bottom).
Middle: The matrices describing the cross-correlations in the noise and mutation-induced deformations of the proteins, which correspond to the dynamics and evolution, respectively. The two correlation matrices share a similar pattern, indicating a correspondence between the dynamics and evolution of protein structures
Credit: Kunihiko Kaneko
In this study, using a database of several thousand proteins belonging to several hundred protein families, the researchers found a strong correlation between protein dynamics (thermal fluctuation) due to thermal noise and structural changes in proteins due to mutations. Furthermore, using a theoretical protein model, they revealed that this dynamism-evolution correspondence is attributable to thermal fluctuation and mutational changes being constrained to a common small-dimensional space. These results also provide a new perspective on the design of functional and artificial intelligence systems.
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




