By comprehensively analyzing the methylation status of approximately 450,000 cytosines on DNA, a research group comprising Assistant Professor Dai Shimizu, Professor Yasuhiro Kodera, and Associate Professor Yusuke Matsui, all from the Nagoya University Graduate School of Medicine; Associate Professor Taniue Kenzui and Associate Professor Nobuyoshi Akimitsu, both from the Isotope Science Center, the University of Tokyo; Associate Professor Hiroshi Haeno from the Graduate School of Frontier Sciences, the University of Tokyo; and Professor Koshi Mimori of Kyushu University Beppu Hospital, Department of Surgery succeeded in producing a methylation panel that can simultaneously diagnose and distinguish between 27 types of malignant tumors. This panel is expected to develop into a screening tool that can accurately identify the location of malignant tumors using liquid biopsy tests, such as blood sampling and urinalysis. The results of this research have been published in the electronic version of Cancer Gene Therapy.
In recent years, tumor-derived ctDNA (circulating tumor DNA) that has leaked into the blood has been attracting attention as a target for identifying malignant tumors. The existence of these tumors can be proved if gene mutations present only in malignant tumors can be detected in the blood through blood sampling. However, most gene mutations found in malignant tumors are common to various tumors. Therefore, even if testing for genetic mutations in the blood reveals the presence of a malignant tumor, it is difficult to identify which organ contains the malignant tumor.
The research group compared and analyzed DNA methylation data from approximately 450,000 locations from a total of 7,950 cases with 28 types of malignancies registered in public databases. By doing so, cytosine characteristically methylated in 27 types of malignant tumors was extracted. Comparison was also performed using normal tissue DNA methylation data from 707 cases, to focus on the abnormal methylation characteristic of malignant tumors, as well as with whole blood DNA methylation data from 95 healthy subjects to be used for diagnosis via blood sampling. Eventually, a methylation panel capable of analyzing 2572 DNA methylation sites was prepared. The panel was clearly able to cluster and distinguish between the 28 types of malignant tumors, including one malignant tumor for which no DNA methylation sites characteristic of cancer tissue could be extracted. Furthermore, for the 27 types of malignant tumors, the case distribution was plotted on a probability density curve based on the β distribution, and the DNA methylation value that minimized false positives and false negatives was set as the threshold for diagnosis of the malignant tumors. From this, 27 types of malignant tumors could be diagnosed with high sensitivity and specificity. The researchers then verified the diagnostic ability of their methylation panel using samples obtained at their research facilities. When the DNA methylation status was examined using a methylation array and the methylation panel was applied to frozen tissue specimens of breast cancer, colon cancer, and gastric cancer, almost all specimens were successfully diagnosed as malignant tumors.
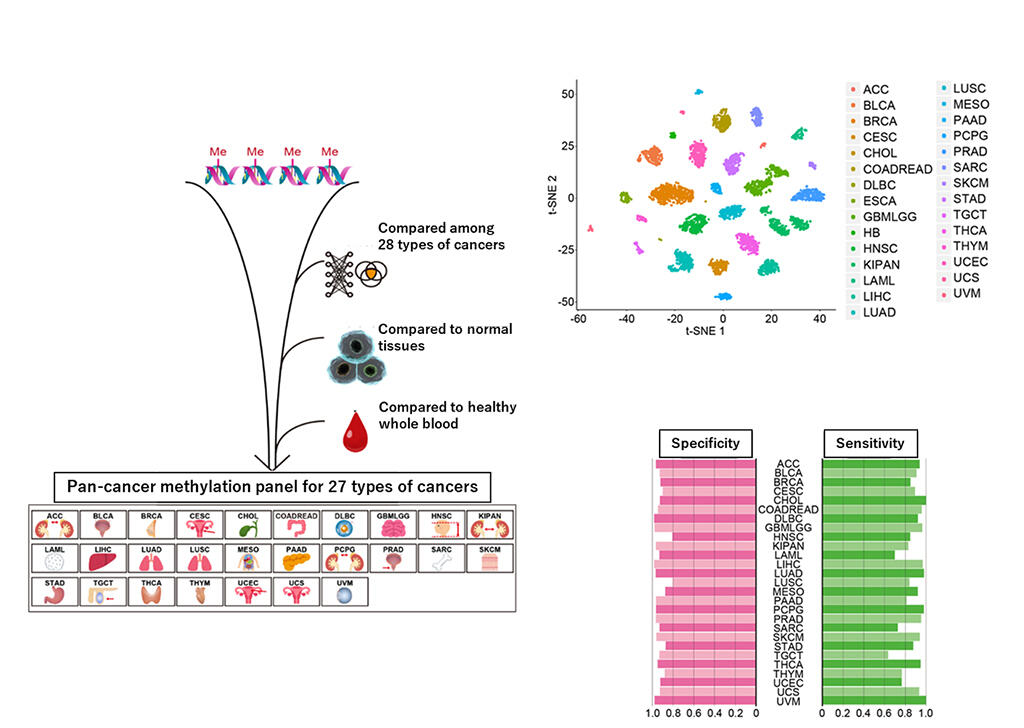
(Provided by Nagoya University)
The public database used to prepare the methylation panel is mainly composed of data from Western countries, but the fact that it was possible to diagnose the tumors in Japanese specimens suggests that the panel can be used universally. Blood sample methylation data from individuals with malignancies from public databases were also obtained and the ability for the panel to be applied to blood sampling tests targeting ctDNA was verified. Upon validation using blood samples of healthy subjects and breast cancer, colon cancer, and lung cancer patients, the false positive rate in healthy subjects was only 3.7%, and breast cancer and colon cancer were identified as corresponding malignant tumors in most cases. On the other hand, in blood samples of lung cancer cases, it was difficult to detect lung cancer because of the small amount of ctDNA in the blood; development of ctDNA methylation analysis technology in the future is expected. In addition, whether a methylation diagnostic panel allows primary lesion estimation in blood data from patients with cancer of unknown primary lesions was verified. The primary lesion identified using the methylation diagnosis panel was largely consistent with the primary lesion suggested considering the clinical course.
The DNA methylation panel developed in this study was able to differentiate between, and diagnose, 27 types of malignant tumors mainly by using data from tumor tissues. Advances in developing ctDNA methylation analysis techniques will allow the application of these approaches in diagnosis via blood tests with a higher accuracy. The research group aims to develop screening techniques that can diagnose up to 27 types of malignancies using a single blood sample.
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




