A group comprised of Associate Professor Kunio Nakatsukasa of the Graduate School of Science, Nagoya City University, Professor Takumi Kamura and Assistant Professor Keisuke Obara of the Graduate School of Science, Nagoya University, and Associate Professor Fumihiko Okumura of Fukuoka Women's University, has discovered a new mechanism by which starved cells fine-tune ribosome biogenesis.
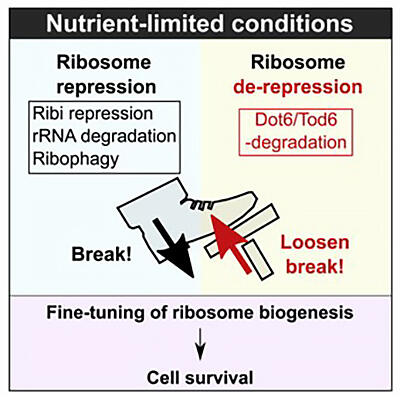
Dot6/Tod6 degradation fine-tunes the repression of ribosome biogenesis under nutrient-limitedconditions
https://doi.org/10.1016/j.isci.2022.103986 CC BY 4.0
Starved cells attempt to conserve energy and prolong their lives by inhibiting energy-consuming reactions as much as possible. Specifically, cell starvation leads to a series of reactions that inhibit ribosome biogenesis through the degradation of ribosomal RNA, suppression of ribosomal protein assembly, ribosome-specific autophagy, and suppression of ribosome biogenesis (Ribi) gene expression. In particular, Dot6 and Tod6 are known as transcriptional repressors involved in the suppression of Ribi gene expression. Thus, it was already known that when cells become starved, or nutrient-limited, it is Dot6 and Tod6 that are activated to suppress Ribi gene expression.
While studying the stability of a variety of proteins within cells, the group noticed that Dot6 and Tod6 were quickly degraded when the cell shifted to a nutrient-limited state. To understand why Dot6 and Tod6 were being degraded despite the fact that they were necessary for the suppression of Ribi gene expression under the nutrient-limited conditions, the group forced the overexpression of Dot6 and Tod6 in the nutrient-limited cells so that the Dot6 and Tod6 were stored rather than degraded. This caused ribosome biogenesis to be overly suppressed, which in turn negatively affected cell growth. This finding demonstrated that the excessive suppression of ribosome biogenesis was not advantageous even when the cell was nutrient-limited, and that it was important for that suppression to be fine-tuned through the degradation of Dot6 and Tod6.
According to Associate Professor Nakatsukasa, "The next question we need to address is whether fine-tuning suppresses all protein biogenesis evenly, or whether it suppresses the biogenesis of a specific protein, or in other words, whether the fine-tuning is quantitative or qualitative in nature. Recent studies have shown that there are ribosomes with slightly different genes, and we are starting to believe that each has a different role. The results of our work may provide clues to understand how these ribosomes with different characteristics are produced, which is an important question to answer in terms of basic biology."
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




