A research group comprised of Lecturer Kaori Yasuda, Research Fellow Toshiyuki Sakaki, and Professor Yasuhiro Isogai of the Department of Pharmaceutical Engineering, Faculty of Engineering, Toyama Prefectural University, and Professor Miki Nakajima and Project Professor Madhu Biyani of the Nano Life Science Institute, Kanazawa University, in collaboration with BioSeeds, has successfully obtained nucleic acid molecules that selectively inhibit the enzymes that inactivate vitamin D.
The group obtained a group of nucleic acid sequences with high affinity for the vitamin D inactivating enzyme CYP24A1 but low affinity for the vitamin D activating enzyme CYP27B1 from a nucleic acid library containing random sequences numbering 10^18. The group analyzed the numerous sequences using next generation sequencing and narrowed the selection down to 11 candidates based on differences in sequence frequency and secondary structure, and then tested those candidates for CYP24A1 inhibition.
The group then analyzed the inhibition effect of four sequences found to inhibit CYP24A1 on other enzymes, and in each case, they found that all four had no inhibitory effect on active vitamin D producing enzymes and sterol metabolizing enzymes with high amino acid homology with CYP24A1, showing that they selectively inhibited CYP24A1.
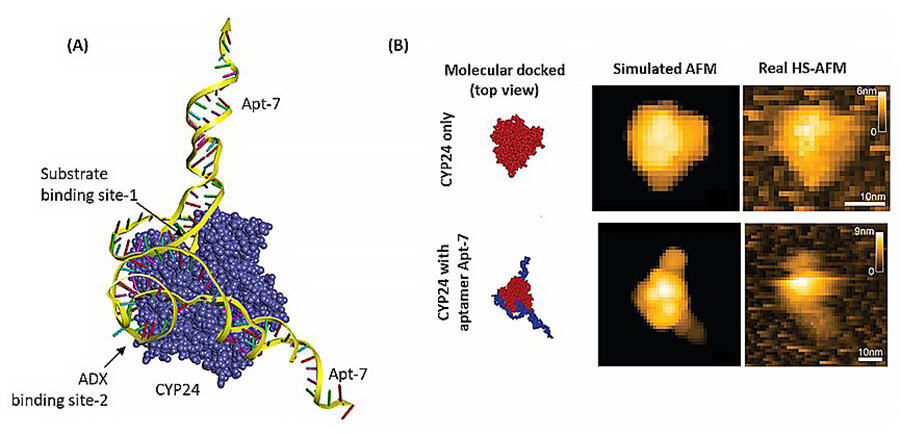
The group followed this by observing the bonding of one of the nucleic acid molecules, Apt-7, with CYP24A1 via high-speed atomic force microscopy. They combined this observation with a molecular docking simulation and enzymological experiment to predict the site of inhibition. Finally, they leveraged a human alveolar basal epithelial adenocarcinoma cell line (A549 cells) to examine the inhibitory effect of Apt-7 on cancer cell proliferation. When comparing the addition of Apt-7 with active vitamin D3 versus the addition of active vitamin D3 alone, the inhibition by CYP24A1 was controlled, increasing the inhibitory effect on cancer cell proliferation.
© 2022 American Chemical Society
According to Dr. Yasuda, "We are currently examining the inhibitory effects of the nucleic acid molecules we obtained on cancer cell proliferation using cell cultures. Moving forward, we will modify those molecules to optimize the sequences and make them more stable within the body while examining the therapeutic effect within the body using mice. In the future, we hope to apply this to treatment to cancers and osteoporosis caused by the over expression of CYP24A1."
Journal Information
Publication: ACS Applied Materials & Interfaces
Title: Novel DNA Aptamer for CYP24A1 Inhibition with Enhanced Antiproliferative Activity in Cancer Cells
DOI: 10.1021/acsami.1c22965
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




