The research group of Professor Koichi Kato of the Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences, and Associate Professor Hirokazu Yagi of the Graduate School of Pharmaceutical Sciences, Nagoya City University, examined proteins that are modified by specific glycans (sugar chains), and succeeded in finding an amino acid sequence that can be called a control code for glycosylations (glycan modifications) which is incorporated into molecular structures. Furthermore, they showed that, by incorporating this molecular code, specific glycosylations can be applied to proteins, opening up new possibilities for use as pharmaceuticals.
Provided by the Institute for Molecular Science
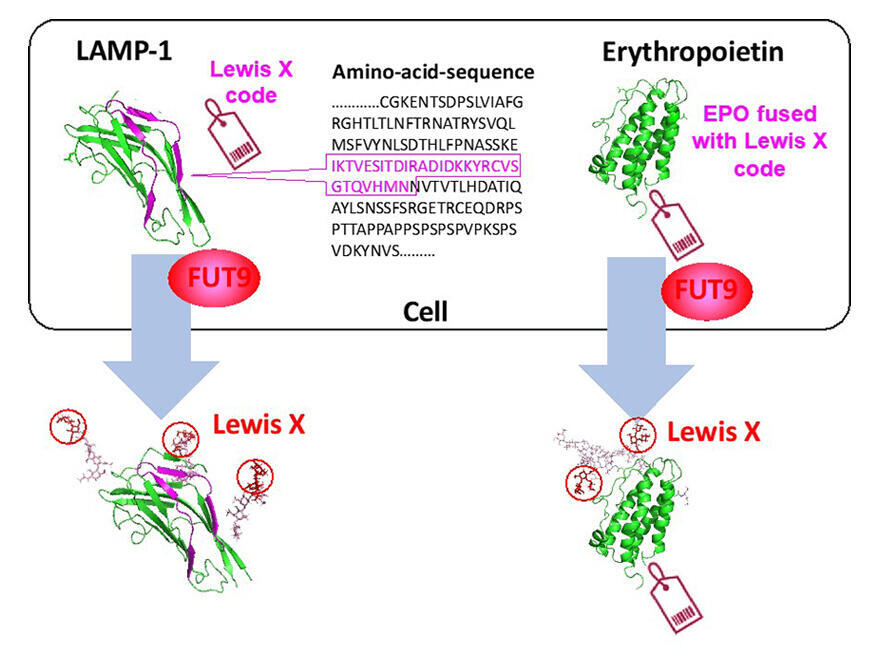
Proteins are created by using genes as blueprints, but there is no clear blueprint for the synthesis of glycans. The research group has been researching on the assumption that part of the blueprint for glycans is hidden in the amino acid sequences (genetic information) of proteins. In their previous research, they revealed that, in mouse neural cells, glycans with a structure called Lewis X specifically modify a protein called LAMP-1. The formation of this Lewis X structure is mediated by fucosyltransferase 9 (FUT9).
In this study, the research group found that the Lewis X modification specific to LAMP-1 occurs not only in neural stem cells, but also several cultured animal cells. LAMP-1 is composed of two domains (N-domain and C-domain) with similar shapes, but, when they were separately expressed in cells, it was found that only the N-domain is modified by Lewis X.
Taking advantage of the similarity of the two domains, they created a series of proteins in which the portions of each domain were interchanged, and, as a result of comparing the glycosylations, determined that the presence of a sequence consisting of 29 amino acid residues in the N-domain caused the Lewis X modification via FUT9 in the surrounding sites. They found that simply linking this sequence to one end of other glycoproteins (to fetuin, a glycoprotein with various physiological effects, and to erythropoietin, which is used as a biopharmaceutical) caused the Lewis X modification in them.
Furthermore, experiments using a proximity-dependent labeling method (a method to mark molecules that are present near the protein of interest) revealed that many erythropoietins with this 29-amino acid segment were present near FUT9. This clarified that the 29-amino acid segment may have a role in facilitating the intracellular encounters with FUT9 in the specific Lewis X modification.
According to Professor Kato, "In the future, there is the possibility that new molecular codes may be successively discovered by studying the various glycoproteins. By using them as regulatory codes for glycosylation, we hope to contribute to the development of biopharmaceuticals and to the advancement of cellular medicine."
Journal Information
Publication: Communications Biology
Title: An embeddable molecular code for Lewis X modification through interaction with fucosyltransferase 9
DOI: 10.1038/s42003-022-03616-1
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




