The human body is made up of around 40 trillion cells, and the nuclei of these cells store approximately two-meter-long genomic DNA, the blueprint for life. Genomic DNA wraps around barrel-shaped histone proteins, forming nucleosomes with an approximately 11-nanometer diameter (nano = one billionth of a meter). Nucleosomes were thought to create a hierarchical structure by folding into a regular helical fiber, but in 2008, Professor Kazuhiro Maeshima of the National Institute of Genetics and his colleagues proposed that there is no regular structures, and that nucleosomes are stored in an irregular and dynamic manner. However, conventional optical microscopes couldn't accurately grasp the very small movements of nucleosomes and their associated DNAs.
Now, a research group consisting of graduate student Shiori Iida of the Graduate University for Advanced Studies, SOKENDAI (SOKENDAI Special Researcher), Professor Maeshima and their colleagues has succeeded in understanding the DNA fluctuations in living nuclei when nuclei in human cells are growing. Because a single cell has the approximately 30 million individual nucleosomes, the group observed the movement of sparsely fluorescently labelled nucleosomes and their associated DNAs using super-resolution fluorescence microscopy. They have also learned that, during the growth process of cell nuclei, DNA fluctuations remain steady. The degree of fluctuation is directly related to the accessibility of DNA by various proteins, therefore cells can read out their genomic information in a similar environment throughout the growth process.
Moreover, the group has discovered that the fluctuations transiently increase when genomic DNA is damaged, suggesting that this facilitates DNA repair because the proteins associated with damage repair can more easily approach the DNA. These research outcomes could be useful in understanding cell abnormalities caused by defects in DNA repair, and associated genetic disorders.
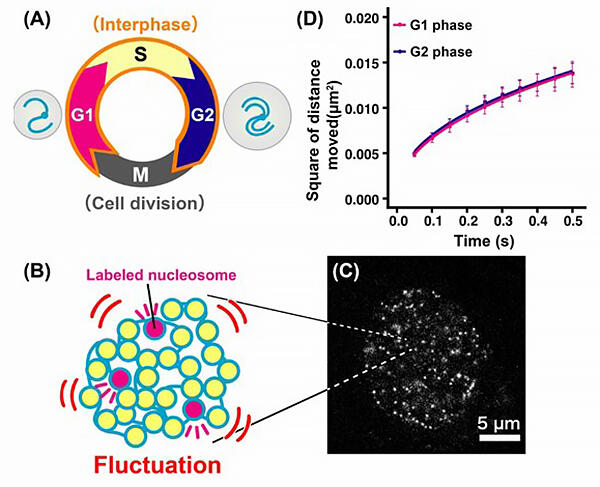
B: To see the movement of the nucleosomes and their associated DNAs, the group labelled a very small fraction of nucleosomes(pink). C: Image of nucleosomes inside the nucleus obtained via super-resolution fluorescence microscopy.
D: In the G2 phase, the quantity of DNA and the size of the nucleus doubled from the G1 phase, but irrespective of this, DNA fluctuations stayed similar throughout the G1, S and G2 phases.




