A research group led by Haruhiko Ehara and team leader Shun-ichi Sekine at the Laboratory for Transcription Structural Biology, RIKEN Center for Biosystems Dynamics Research has revealed that RNA Polymerase II (RNAP II), which is responsible for gene expression in eukaryotic life, maintains chromatin structures by reassembling once-unwound nucleosomes after transcription. They collaborated with Research Associate Tomoya Kujirai and Professor Hitoshi Kurumizaka from the Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, the University of Tokyo. The group used a cryogenic electron microscope (cryo EM) to capture the molecular structure of these processes as if performing time-lapse photography. It's hoped the findings will assist in the development of treatments for diseases caused by transcriptional control and the breakdown of these processes. The results were published in the August 18 issue of the international scientific journal Science.
Provided by RIKEN
In eukaryotic cells, genomic DNA is compactly stored and protected in the nucleus through a chromatin structure. This structure consists of a beaded structure of basic units called nucleosomes, in which DNA is wrapped twice around a core-like structure made of eight histone proteins (histone octamer).
It was previously known that DNA transcription is performed by RNAP II (an enzyme), which follows DNA from upstream to downstream. As a fundamental rule, DNA must be unwound once transcription is carried out with the chromatin structure maintained. However, how RNAP II performs transcription in this configuration has long been a mystery.
So far, the research group has successfully constructed an experimental system in which chromatin is reformed in vitro and transcription is artificially reproduced. The way in which RNAP II proceeds by peeling off DNA from histones in stages was revealed between 2017 and 2019.
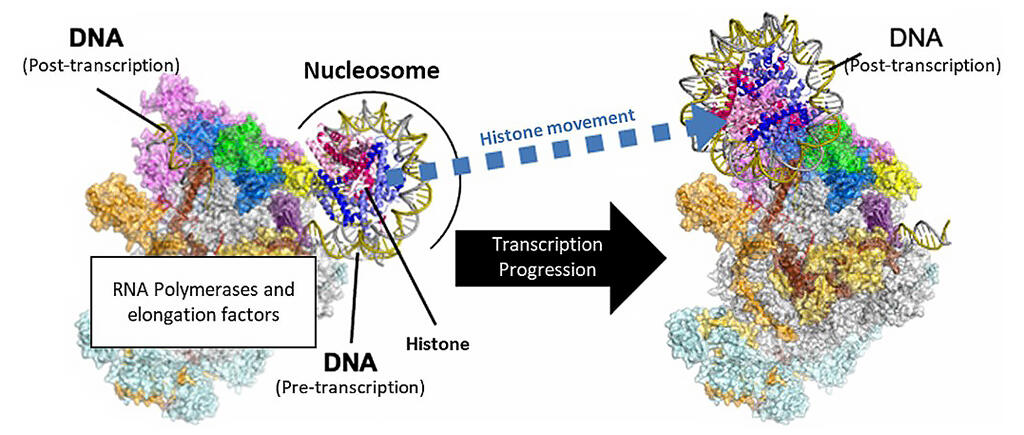
The group used their experimental system to induce RNAP II to transcribe nucleosomes, analyzing the structure of the composite formed in the process of RNAP II passing through the central part (dyad) of nucleosomal DNA for one nucleosome using a cryo EM.
Specifically, the nucleosomal DNA is designed to stop at four positions between the start and end of transcription. In addition, six primary transcription elongation factors and a histone chaperone (FACT), which were reported to be required for transcription in cells, were synthesized and transcribed in their presence.
Cryo EM analysis of each of the composites formed yielded six different complex structures with stops at the 42nd (near dyad), 49th (just before dyad), 58th (just after dyad), and 115th (nucleosome exit) base pair positions from the entrance where RNAP II enters the nucleosome. The group also revealed for the first time that the composite forms a giant transcription factor complex (EC), in which transcription elongation factors bind to RNAP II and cover the surface except for the DNA entry and exit sites.
By obtaining a tiered molecular structure, the group succeeded in capturing how the EC gradually unwinds the DNA from the histone, and at the point of dyad passage, FACT binds and transfers the histone to the upstream side, where transcription ends. FACT bound to histones just before the dyad when DNA was detached from histones, bound to hexamer and octamer histones just after the dyad, and detached at the nucleosome exit.
Sekine commented, "In our study, we focused on histone chaperones and transcription elongation factors, but many other factors act on RNAP II and nucleosomes. In the future, we want to conduct research to understand transcription and chromatin function involving more factors. We believe this will eventually lead to a better understanding of diseases caused by the breakdown of transcriptional control."
Journal Information
Publication: Science
Title: Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT
DOI: 10.1126/science.abp9466
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




