A research team made up of Specially Appointed Researcher Yuji Okamoto (now Specific Assistant Professor of the Graduate School of Medicine, Kyoto University), Research Associate Masashi K. Kajita (Currently Assistant Professor of the Faculty of Engineering, University of Fukui) and Professor Kazuyuki Aihara (now University Professor of the University of Tokyo) of the Institute of Industrial Science, the University of Tokyo, Professor Arinobu Tojo of the Institute of Medical Science, the University of Tokyo (now Executive Director/Executive Vice President of Collaboration, Data Science and Faculty Affairs at Tokyo Medical and Dental University) and their colleagues has developed a mathematical model that can quickly predict the therapeutic outcomes of nilotinib, a therapeutic drug for chronic myeloid leukemia (CML), for each patient. Their outcomes were published in npj Systems Biology and Applications.
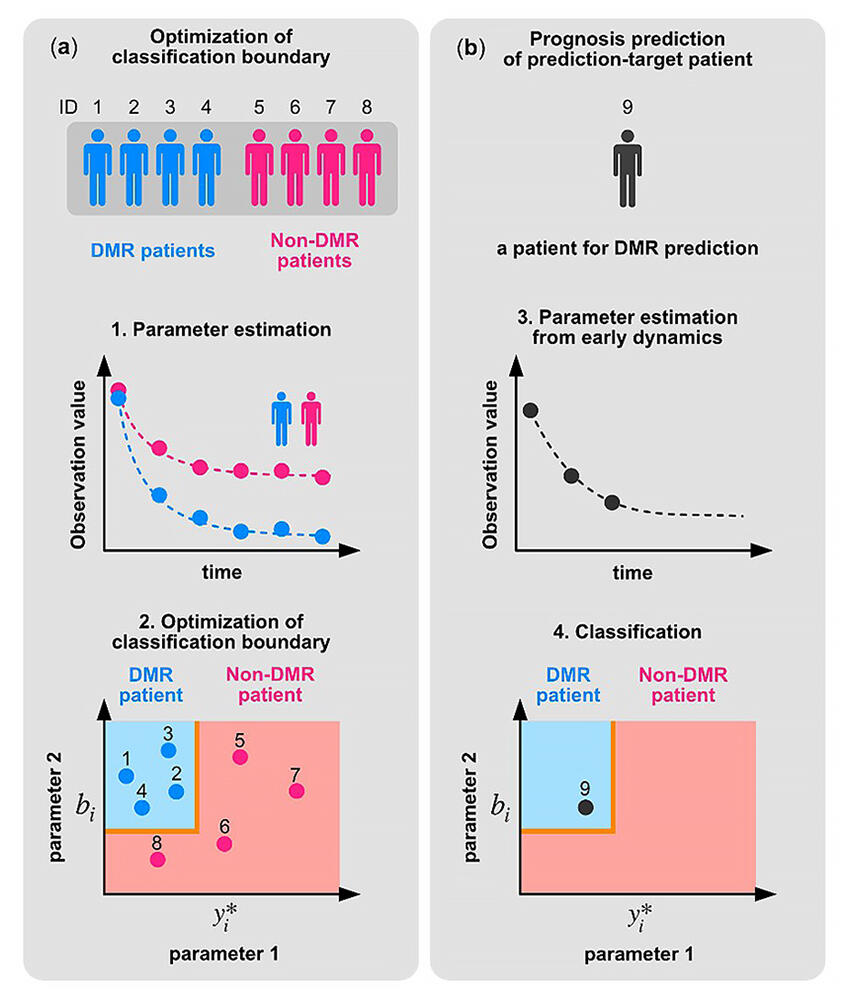
(a) Learning steps. The model parameters of each patient are estimated using the time series data and mathematical model of CML dynamics (Step 1). In the estimated parameter space, the boundary between the patients who achieved deep molecular response (DMR patients) within two years and the patients who did not achieve DMR (non-DMR patients) is obtained by optimization (Step 2).
(b) Prediction steps. The parameters of a prediction-target patient are estimated from the initial time series data up to 6 months after the start of treatment (Step 3). The parameters of the prediction-target patient are plotted on the classification boundary determined in Step 2. The patients whose parameters are in the blue area are predicted to achieve DMR within two years. In this example, the prediction-target patient is predicted to achieve DMR (Step 4).
Provided by the University of Fukui
Chronic myeloid leukemia (CML) is caused by a mutation of the hematopoietic stem cells that are the source of white blood cells, red blood cells, platelets and more. The leukemia cells (CML cells) that can be differentiated from mutant hematopoietic stem cells have the BCR-ABL1 fusion gene formed by reciprocal translocation between two chromosomes. The disordered propagation of CML cells occurs due to the abnormal activation of the tyrosine kinase enzyme created by this gene.
At the moment, therapeutic drugs known as BCR-ABL1 tyrosine kinase inhibitors (TKI) are available, and these can cause a great reduction in CML cells. They improve the prognosis of large numbers of patients, but there are individual differences in their effects.
Their therapeutic effect on CML is shown using an international scale (IS value). The IS value reflects the expression level of the BCR-ABL1 fusion gene, and is measured via blood tests. If, in accordance with treatment, the CML cell level drops sufficiently and a deep molecular response (DMR) occurs, the IS value will also become sufficiently small.
At the moment, the TKI given to newly diagnosed CML patients consists of the first-generation TKI imatinib, and the second-generation dasatinib, nilotinib and bosutinib. The medical treatment options consist of switching to a different TKI if the effects of the TKI administered are poor. To do this, doctors need to quickly predict the effects of TKI treatment for each patient.
The research group has proposed a mathematical model that can predict the efficacy of nilotinib for each CML patient from time series data over a short time period. More specifically, the group has created a classification method that can estimate whether a CML patient will reach DMR within two years of nilotinib administration, based on the total amount of white blood cells in their peripheral blood and the IS value when nilotinib is first administered, and in the third and sixth months after administration begins. The key point is the focus on temporal changes in the CML cell level in peripheral blood.
First, the group expressed the temporal changes in the levels of CML cells and regular white blood cells in the peripheral blood using a simple ordinary differential equation model made from four parameters. By altering these parameters for each patient, it was possible for the group to simulate the temporal changes in the levels of CML cells and regular white blood cells in the peripheral blood for each patient. However, it is not possible to differentiate and measure CML cells and regular white blood cells in normal blood tests. Thus, the group developed a mathematical model that estimates the levels of CML cells and regular white blood cells in the peripheral blood, which cannot be measured directly, from the measurable IS value and the total level of white blood cells in the peripheral blood.
Next, using data from a clinical trial with CML patients, the group estimated the parameters for an ordinary differential equation model associated with the level of CML cells from two years' worth of time series data consisting of the total amount of white blood cells in peripheral blood and the IS value of each CML patient. From this, they learned that patients can be categorized into those who reached DMR and those who did not, based on these estimated parameters.
They also split patients into a classification model learning group and a response prediction group. First, they determined the optimal classification boundary, based on whether DMR was reached and the individual patient parameters estimated from two years' worth of time series data belonging to the learning group of patients. Then, they used the data of the group up to the sixth month to estimate the parameter values for those patients and predicted whether they reached DMR based on the classification boundary. As a result, they were able to predict whether a patient would reach DMR with a high level of accuracy - approximately 94% - when using their optimized method to determine the classification boundary, based on a standard DMR known as MR4.5.
The clinical trial data used for this study consisted of data from 32 patients, and the group only verified efficacy with nilotinib, so in the future, an investigation into the efficacy of diverse TKI using large amounts of clinical data is required. However, showing that it is possible to predict the effects of a TKI for each patient using blood tests has major significance.
Journal Information
Publication: npj Systems Biology and Applications
Title: Early dynamics of chronic myeloid leukemia on nilotinib predicts deep molecular response
DOI: 10.1038/s41540-022-00248-3
This article has been translated by JST with permission from The Science News Ltd.(https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




