A research group led by Project Assistant Professors Chihong Song and Kazuyoshi Murata of the National Institute for Physiological Sciences, Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences, in collaboration with a research group led by Professor Kaoru Mitsuoka of the Research Center for Ultra-High Voltage Electron Microscopy, Osaka University, has successfully revealed the particle structure of the tokyovirus, a type of giant virus, at a resolution of 7.7 Å using an ultra-high voltage cryo-EM installed at the Center for Ultra-High Voltage Electron Microscopy.
Provided by NINS
Giant viruses are viruses that are large enough to rival small bacteria in both their physical and genomic size. However, their size is a factor that makes structural studies of giant viruses difficult. In the structural analysis of a typical virus particle, a medium-sized transmission electron microscope with an acceleration voltage of electrons up to 300 kV is used to reconstruct its three-dimensional structure from the numerous projected images of the virus. This method is limited to a few hundred nanometres in which the electron beam penetrates the sample sufficiently and is in focus at one time, and accurate projection images cannot be recorded for giant viruses that are beyond this range. Until now, it has been necessary to analyze the partial structure around the focus area and reconstruct the structure of an entire giant virus by piecing it together.
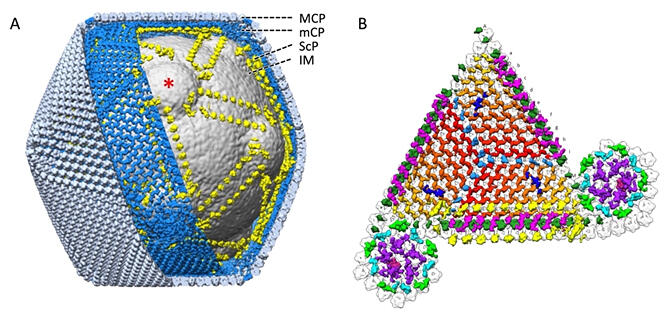
By recording high-resolution projection images of the tokyovirus, which has a diameter of 250 nm, the research group was able to reveal its entire structure at a resolution of 7.7 Å using ultra-high voltage cryo-electron microscopy at an acceleration voltage of 1 MV. This revealed the detailed structure of a novel giant capsule for storing giant viral genes.
The surface of many virus particles are covered with a common, regularly arranged major capsid protein (MCP). Tokyoviruses possess a novel network structure of minor capsid proteins (mCPs) that supported this directly under the MCPs. It was also found that a scaffolding protein (ScP) developed between the mCP and the nuclear membrane that envelopes the giant viral gene, which functions to form the protruding structure on the nuclear membrane that is characteristic of this giant virus. More unknown proteins were located outside of the MCP. A close relative of the tokyovirus is known to adhere to the surface of host amoeba. This suggested that the protein at this tip may be involved in adherence to the host.
"Giant viruses are so large that their particle size and gene size are comparable to those of small bacteria," explained Murata. "They are too large to be observed with a conventional electron microscope and too small to be observed with an optical microscope. In 2016, we precisely imaged the entire structure of the tokyovirus found in the Arakawa River in Tokyo. We hope to reveal more giant virus structures using ultra-high pressure cryo-electron microscopy."
Journal Information
Publication: Scientific Reports
Title: A novel capsid protein network allows the characteristic internal membrane structure of Marseilleviridae giant viruses
DOI: 10.1038/s41598-022-24651-2
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




