A research group led by Associate Professor Wataru Yoshida of the School of Bioscience and Biotechnology, Tokyo University of Technology, and Professor Ryutaro Asano of the Institute of Engineering, Tokyo University of Agriculture and Technology, has developed a method for constructing luminescent proteins that can easily detect various modified bases in genomic DNA, which is anticipated to be used as a biomarker for cancer and other diseases.
Provided by Tokyo University of Technology
DNA methylation is a reaction in which a methyl group is added at the fifth position of cytosine in a contiguous sequence of cytosine and guanine (CpG), mainly in genomic DNA, and is involved in the regulation of gene expression. The continuous oxidation of 5‐methylcytosine produces 5‐hydroxymethylcytosine, 5‐formylcytosine and 5‐carboxycytosine.
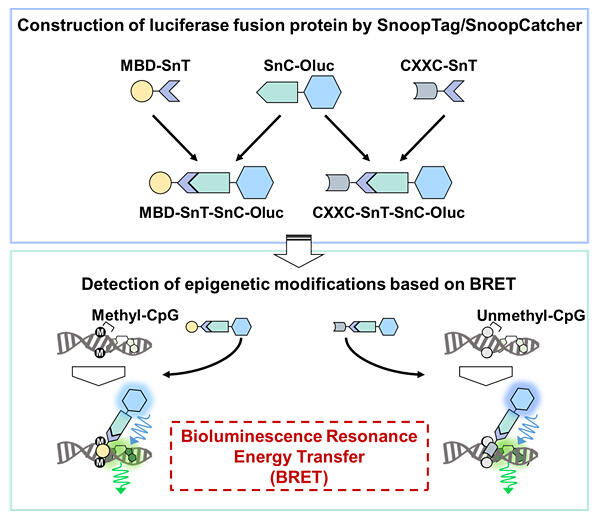
The research group has previously fused luciferases to methyl CpG‐binding domains (MBDs) and non‐methyl CpG‐binding domains (CXXCs) and developed methods to measure these modified bases using bioluminescence resonance energy transfer. Based on these results, the researchers considered that a simple method for constructing a fusion protein between the target modified base recognition protein and luciferase would make it possible to develop a more convenient method to measure modified bases.
They focused on the protein ligation system SnoopTag (SnT)/SnoopCatcher (SnC) as a method for constructing fusion proteins. SnT and SnC spontaneously ligate simply by standing at room temperature. Various proteins were produced recombinantly by fusing SnT to MBD (MBD‐SnT), SnT to CXXC (CXXC‐SnT) and luciferase to SnC (SnC‐Luc). The researchers observed spontaneous linkage when each of these was mixed and allowed to stand for one hour at room temperature. Furthermore, using these ligation products, they found that the methyl CpG and non‐methyl CpG content of genomic DNA can be measured using bioluminescence resonance energy transfer.
"Using this method, it is possible to construct fusion proteins of modified base‐recognition proteins and luminescent proteins in arbitrary combinations," commented Yoshida. "By fusing luciferases with different luminescent properties to various modified base‐recognition proteins, we hope to be able to develop a method to measure these modified bases simultaneously."
Journal Information
Publication: Analytical Chemistry
Title: Universal Design of Luciferase Fusion Proteins for Epigenetic Modifications Detection Based on Bioluminescence Resonance Energy Transfer
DOI: 10.1021/acs.analchem.2c05066
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




