The research group of Professor Koichi Kawakami (Laboratory of Molecular and Developmental Biology, National Institute of Genetics (NIG)) and Dr. Hideo Mori (Osaka Habikino Medical Center) identified a mutant strain of SARS‐CoV‐2 that does not show positive results using conventional PCR. The result was published in the international scientific journal Cureus.
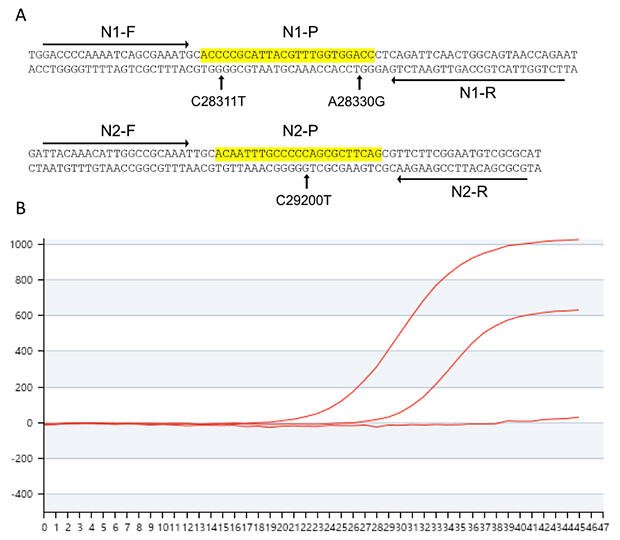
B. Amplification curves of omicron variants by RT‐qPCR using N1 and N2 primer‐probe sets.
Provided by NIG
Reverse transcription quantitative polymerase chain reactions (RT‐qPCR) are used to diagnose infections with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which cause COVID‐19 symptoms. For RT‐qPCR examination, the primer/probe sequences targeting two regions of the N gene from SARS‐CoV‐2 are used in RT‐qPCR kits manufactured by Takara Bio and TOYOBO, among others. Since the pandemic began, many sub strains of SARS‐CoV‐2 containing gene mutations have emerged. The Omicron sub strain is currently widely disseminated worldwide, including in Japan.
In mid‐October of 2022, a saliva specimen collected from a patient presenting with a fever in Osaka showed positive results using multiple different nucleic‐acid tests; however, the RT‐qPCR kit using N1 and N2 primer probes could not detect SARS‐CoV‐2. The research group then performed a detailed analysis of the sample.
Analysis of the S‐gene (3) region in the saliva specimens revealed that the infected strain was an Omicron sub strain. The researchers predicted that this sub strain has an unknown mutation in the N gene and examined the detailed nucleotide sequences of the N1 and N2 regions. The results revealed two mutations in the N1 region (C28311T and A28330G) and one mutation in the N2 region (C29200T). These mutations appear to be undetectable using conventional primer‐based RT‐qPCR test kits. Another known omicron strain, BA5.2, has two mutations in the N1 region (C28311T and A28330G) but no mutation in the N2 region.
The RT‐qPCR kit manufactured by Takara Bio detected the PCR amplification product in BA5.2, although its levels was approximately half of the amount in the positive control; in contrast, the PCR amplification product was not detectable in the present sample.
Strains containing three mutations in the N1 and N2 regions showed frequencies of 0.23% and 0.06% of all virus sequences in Japan and worldwide, respectively. These values may be underestimated because the viruses containing these three mutations were not detected using some test kits. Due to this, PCR testing with N1 and N2 primer probes may have missed some patients with COVID‐19. Even if the PCR test using conventional primers does not give a positive result, mutant strains can be detected if PCR is performed using primers‐probes for different locations after screening patients with characteristics symptoms of COVID‐19.
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




