Since the year 2000, the incidence of nontuberculous mycobacteria (NTM) pulmonary disease has increased dramatically in Japan, surpassing tuberculosis. NTM is the generic name given to mycobacteria other than the Mycobacterium tuberculosis complex and Mycobacterium leprae. As treatment strategies differ depending on the species and subspecies, accurate identification of NTM at the subspecies level is necessary for appropriate treatment.
A research group led by full‐time Specially Appointed Assistant Professor Kiyoharu Fukushima of the Laboratory of Host Defense at the World Premier Institute Immunology Frontier Research Center, Osaka University (IFReC), Specially Appointed Assistant Professor Yuki Matsumoto and Associate Professor Shota Nakamura at the Department of Infection Metagenomics of the Genome Information Research Center, Research Institute for Microbial Diseases (RIMD), Osaka University developed a new method for NTM diagnosis using cloud computing in collaboration with Director Hiroshi Kida of the Department of Respiratory Medicine at Osaka Toneyama Medical Center. This study was published in the Journal of Clinical Microbiology.
Provided by the Research Institute for Microbial Diseases
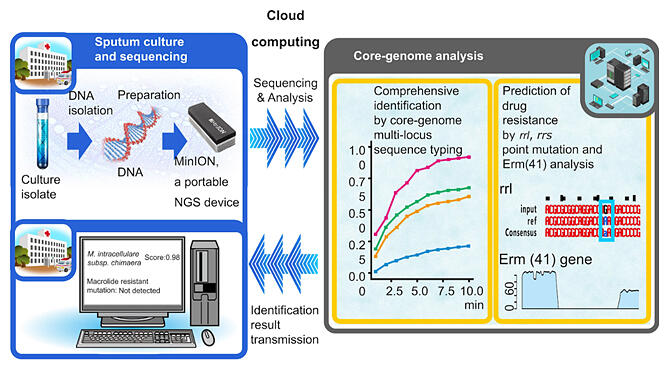
Diagnosis of NTM pulmonary disease is expensive and time‐consuming, requiring 2‐8 weeks for the culture test and more than 1 month for subspecies identification and drug susceptibility testing. By combining a large‐scale genome database comprehensively covering 175 NTM species and a drug susceptibility prediction algorithm based on known drug‐resistant genes, the research group established a method for identifying subspecies and predicting drug susceptibility in cultured samples within a maximum of 3 days using nanopore sequencing and core genome analysis. They also established a system to receive the results in a timely manner, even at remote medical institutions, through cloud computing. Simple gene sequencing is performed in a hospital laboratory, and the results are sent to the cloud of RIMD, where possible strains can be identified in approximately 10 minutes.
A prospective investigation of 116 patients with NTM that were newly or previously diagnosed in Osaka Toneyama Medical Center showed 99.1% accuracy in species‐level identification and 84.5% in subspecies‐level identification. Resistance to antimicrobics of macrolide and amikacin was detected with specificities of 97.6% and 100%, respectively. They also used the system to analyze a patient with NTM pulmonary disease in whom an unidentifiable strain was repeatedly detected, leading to the discovery of three new species.
Fukushima commented on the outcome, "In this study, 20% of patients were macrolide resistant, and we feel that resistant bacteria are spreading. We are currently consulting with the Ministry of Health, Labour and Welfare (MHLW) to ensure that this method can be used in actual medical practice."
Journal Information
Publication: Journal of Clinical Microbiology
Title: MGIT-seq for the Identification of Nontuberculous Mycobacteria and Drug Resistance: a Prospective Study
DOI: 10.1128/jcm.01626-22
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




