Until now, there has been no database on membrane permeability, which would be an important tool for cyclic peptide drug discovery. There have been calls for the creation of a specialized database because of the difficulty in comparing values obtained from different measurement methods. From this background, a group led by Professor Yutaka Akiyama, Assistant Professor Keisuke Yanagisawa and graduate student Jianan Li of the School of Computing at Tokyo Institute of Technology has created the CycPeptMPDB database (Cyclic Peptide Membrane Permeability Database), which comprehensively collects and provides free access to experimental data on the membrane permeability of cyclic peptides, from literature and other sources.
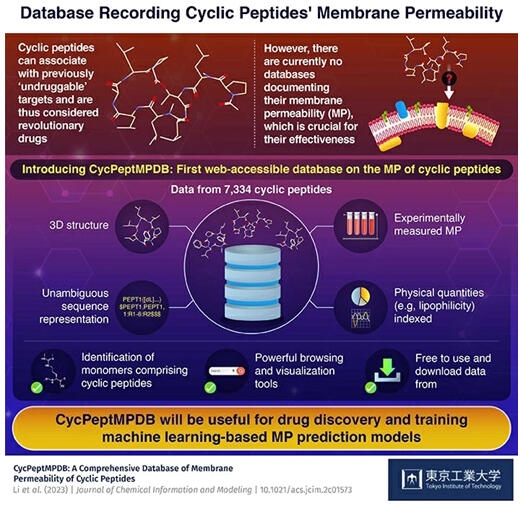
The database contains membrane permeability data on 7334 cyclic peptides from papers and patents, as well as their chemical structures and three‐dimensional conformational data in a unified format. The database is expected to lead to efficient cyclic peptide drug discovery. The results were published in the online version of the Journal of Chemical Information and Modeling.
Peptide drug discovery has attracted attention as an alternative to small molecule drug discovery. Particularly, many researchers and companies are working on the discovery of cyclic peptide drugs, which can improve stability in the body compared to linear peptides. Japan has also been developing technologies for many years to rapidly and automatically find cyclic peptides with high binding affinity to their targets. On the other hand, simply having a high binding affinity to a target is insufficient for use as a drug. It should exist stably in the body without side effects and reach the site where the target exists.
Under such conditions, the most important issue in cyclic peptide drug discovery is cell membrane permeability, which is necessary for the drug to reach the interior of the cell. Cells are surrounded by a thin lipid membrane (lipid bilayer membrane), and if a drug is designed to target a protein or other substance inside the cell, it must efficiently pass through this membrane. Cyclic peptides are larger in size than traditional small molecule drugs and generally have lower membrane permeability.
However, in the natural world, not all cyclic peptides are incapable of passing through the cell membrane. Some cyclic peptides are highly membrane‐permeable. Even when cyclic peptide drugs with excellent binding affinity have been developed, it has been difficult to increase membrane permeability with subsequent minor modifications, and in many cases drug discovery has come to a standstill.
To solve this problem, research has been conducted worldwide to elucidate the principles that allow an increase in membrane permeability. However, the experimental data are scattered across numerous references, and data collection has been a major barrier to new researchers entering the field.
The CycPeptMPDB developed by the group is based on a comprehensive review of more than 40 recent papers and patents. Cyclic peptide membrane permeability data were collected and classified, and the results of an advanced information analysis were added to this database. Currently, 7334 cyclic peptide membrane permeability measurements are available, with detailed links to the experimental methods used and publication groups, allowing the easy retrieval of data sets that meet specific criteria.
Cyclic peptides have complex molecular structures, but CycPeptMPDB uses a flexible notation system called the hierarchical editing language for macromolecules (HELM) format to represent all the collected cyclic peptide structures in a consistent manner. For user convenience, the chemical structures are also described in the simplified molecular input line entry system (SMILES) format, and representative examples of the three‐dimensional structures (stereoconformations) are adopted by each cyclic peptide molecule were also calculated and included in the database. It also has a visualization function for various statistics showing the data distribution and others. The data can also be downloaded in batches.
This research group has developed various technologies related to cyclic peptides. The first author, Jianan Li, a graduate student, has already developed and published a paper on a system that predicts the persistence of cyclic peptides in the body using deep learning technology, a type of AI. At first glance, it seems that a similar approach could be used to predict membrane permeability, but in the case of membrane permeability prediction, there are many caveats such as the inability to easily compare measurements owing to differences in the measurement methods. Additionally, the phenomenon of membrane permeability itself is much more complex and requires more data collection, and with greater care, compared with persistence prediction in the body.
The CycPeptMPDB has been developed with these points in mind and based on the know‐how of organization methods that have been developed to meet research needs. In addition to regularly increasing the number of entries, the accuracy of the information analysis provided (e.g., the presentation of the three‐dimensional conformation of cyclic peptides and calculation of the surface area) will be improved in the future. Moreover, because the method for predicting the membrane permeability of cyclic peptides using advanced physicochemical simulation technology requires a huge amount of computation time, efforts will also be made to develop an approach to easily predict membrane permeability using AI based on various characteristics related to how they move in performing very short simulations only.
Journal Information
Publication: Journal of Chemical Information and Modeling
Title: CycPeptMPDB: A Comprehensive Database of Membrane Permeability of Cyclic Peptides
DOI: 10.1021/acs.jcim.2c01573
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




