The research team led by Associate Professor Tetsuya Kitaguchi of the Institute of Innovative Research at Tokyo Institute of Technology and Graduate Student Yoshihiro Ito of the School of Life Science and Technology at the same institute, in collaboration with Ajinomoto Co., Inc., developed a novel high-throughput method for screening microbial strains, thereby enabling the rapid production and secretion of biopharmaceuticals. The results were published in the online journal Small.
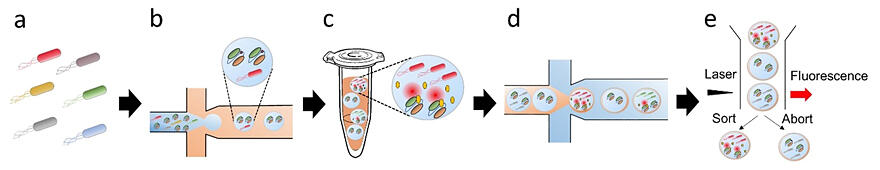
(a, b) A solution containing target protein-secreting microorganisms and Q-body is poured into a microfluidic channel and sheared with oil to prepare a water-in-oil emulsion.
(c) Microorganisms are cultured in the emulsion, and the target protein is detected as fluorescence by a secretory production biosensor.
(d) To prevent emulsion breakage during sorting, emulsions are passed through microfluidic channels to prepare water-in-oil-in-water emulsions protected by an oil layer.
(e) Emulsions in which target proteins are secreted are sorted and collected using fluorescence intensity as an indicator.
Provided by Tokyo Institute of Technology
In the production of biopharmaceuticals, the use of microorganisms for producing and secreting proteins is widely applied because of the low cost of the technique and avoidance of contamination by animal-derived components. However, the generation of industrial microbial strains for such purpose requires the introduction of many genetic modifications to achieve optimal protein production and secretion, and the long development period is an issue.
The research group focused on massively parallel culture with a culture reactor and used a water-in-oil-in-water (W/O/W) emulsion (a dispersion of microdroplets) to grow and evaluate a large-scale library of genetically modified microbial strains within a short period. They managed to encapsulate genetically modified microbial strains one by one within microdroplets (∼30 micrometers in diameter) by carrying out both the bacterial cell growth and target protein production and secretion processes in the emulsion. Then, they screened and selected the strains that showed improved protein production and secretion abilities.
A quenching antibody (Q-body) was used to detect the target protein secreted into the emulsion. Q-body is a fluorescently labeled antibody fragment. When it binds to an antigen protein, the fluorescence intensity increases rapidly in an antigen concentration-dependent manner, making it a promising biosensor for emulsion culture.
First, the team verified whether it was possible to detect changes in the target protein concentration using Q-body in the emulsion. To test it as a biopharmaceutical model for detecting human fibroblast growth factor 9 (FGF9), various concentrations of this growth factor fused with a peptide sequence that responds to Q-body were first prepared. Then, the samples were encapsulated within the emulsion droplets together with the fluorescent scFv-type Q-body modified with the TAMRA dye, and the fluorescence intensity was analyzed using flow cytometry.
A concentration-dependent increase in fluorescence intensity was observed at FGF9 molar concentrations in the range of 10 to 1000 nmol, indicating that protein detection using Q-body in the emulsion was possible.
Next, the gram-positive bacterium Corynebacterium glutamicum, which produces and secretes FGF9, and a negative control strain that does not secrete this protein were separately encapsulated and cultured, and the possibility of selective recovery of the FGF9-producing strain alone was examined using the fluorescence detection method. The team found that the emulsion with the FGF9-producing strain showed significantly higher fluorescence intensity than the emulsion with the negative control, and the selective recovery of only the producer strain using the fluorescent Q-body detection method was successful.
After constructing a library of more than 107 FGF9-producing mutant strains generated with the chemical mutagen N-methyl-N′-nitro-N-nitrosoguanidine, the research group cultured 105 of those strains in the W/O/W emulsion and demonstrated that the top 50 strains showing high fluorescence intensity could be selectively recovered with a cell sorter. Further culture and evaluation studies revealed that the secretory production of FGF9 by the screened mutant strains was significantly improved (∼3 times increased amount) compared with that by the non-mutated strain.
These findings demonstrate the possibility of rapid strain selection from a large-scale library-which is impossible with existing methods-as well as the utility of the emulsion culture and Q-body method for screening strains with high protein production and secretion capabilities.
The developed method enables the rapid screening of industrial microbial strains used for the mass production of biopharmaceuticals, the demand for which has increased in recent years. Its additional benefit is the low-cost supply of various biological drugs within a few years. In the future, the research group will apply this screening technique in the research and development of biopharmaceuticals and extend its application to useful proteins other than biological medicines. Additionally, they will improve the efficiency and accuracy of the screening method to achieve even higher precision by applying Q-body with Förster resonance energy transfer.
Journal Information
Publication: Small
Title: Efficient Microfluidic Screening Method Using a Fluorescent Immunosensor for Recombinant Protein Secretions
DOI: 10.1002/smll.202207943
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




