The research group led by Junior Associate Professor Morito Kurata and Graduate Student Keisuke Sugita of the Graduate School of Medical and Dental Sciences (Medicine) at Tokyo Medical and Dental University (TMDU) has developed a new screening method to clarify the mechanism of the cancer microenvironment and successfully identified the key factors of drug resistance with cell−cell interactions. The new screening method is expected to be applicable to the development of new drug targets for treatment-resistant tumors such as leukemia and pancreatic cancer.
Provided by TMDU
In cancer therapy, clarification of the drug resistance mechanism that makes anticancer drugs ineffective to tumor cells is an important issue to be overcome. It is known that the 'tumor microenvironment,' constructed by cancer cells, cancer stem cells, and other stromal cells, also affects drug resistance, and this has become a major research topic.
Many reports on the conventional 'cancer microenvironment,' however, have focused on cancer itself, with poor prognosis. In contrast, Kurata's team worked to establish a new clustered regularly interspaced short palindromic repeats (CRISPR) screening method to comprehensively understand the drug resistance mechanism induced by peritumoral supporting cells.
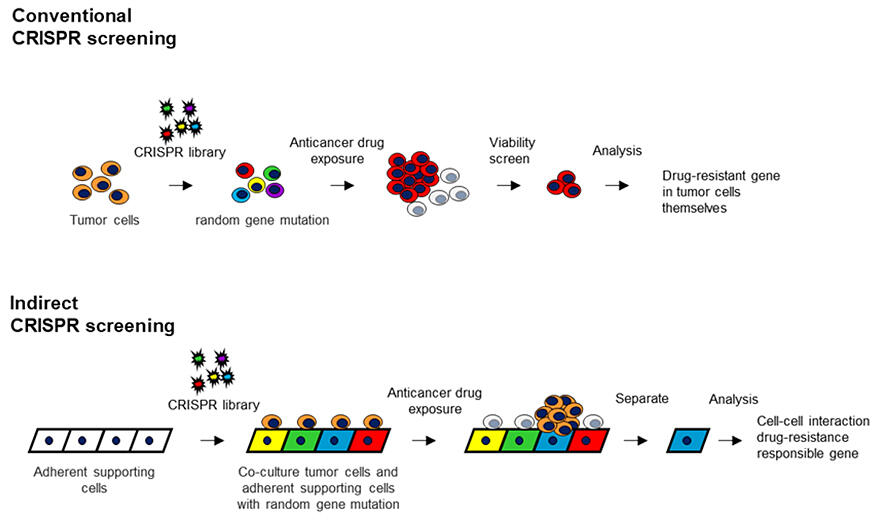
For the identification of the candidate genes in drug resistance, conventional CRISPR screening employs CRISPR libraries that induce random mutations, and selective survival and death of tumor cells are then detected under conditions such as drug exposure. However, when drug resistance arises from cell−cell interactions in the surrounding environment such as the 'cancer microenvironment,' it has been difficult to identify candidate genes responsible for drug resistance through cell−cell interactions.
This is because even if random mutations are induced in the surrounding supporting cells, the supporting cells themselves do not selectively survive or die under drug exposure. The research team then devised a new method that combine a system to isolate supporting cells that can induce drug resistance and named it 'indirect CRISPR screening.'
In this study, Dendra2, a protein whose fluorescent wavelength changes under ultraviolet light, was first incorporated into supporting cells, and when random mutations induce a state that tumor cells can survive the drug, drug-resistant inducible cells are separated from the cells that could not induce drug resistance. From this, the cells responsible for the drug resistance are analyzed by irradiating ultraviolet light into the supporting cells which anchor tumor cells responsible for drug resistance, and by converting the fluorescence wavelength from green to red.
The research team identified the candidate genes from the cells recovered using this method. Then, to confirm whether cell death could be suppressed by anticancer drugs for the obtained candidate genes, they induced deletion of each candidate gene in supporting cells once again and conducted co-culture experiments with tumor cells. The results showed that C9orf89, MAGI2, MLPH, and RHBDD2 suppressed cell death among the candidate genes.
The researchers further analyzed a pancreatic cancer specimen bearing abundant stromal cells around the treatment-resistant tumor cells and found in the candidate gene RHBDD2that the negative group had a significantly shorter overall survival period than the positive group.
With these results, the research team considers the possibility that RHBDD2, which was identified in the new screening method, is deficiently expressed in the stroma around the tumor, thereby inducing resistance to anticancer drugs and shortening survival period.
This research was funded by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT). The research results were published on June 1st in the online edition of the international scientific journal Communications Biology.
Journal Information
Publication: Communications Biology
Title: Indirect CRISPR screening with photoconversion revealed key factors of drug resistance with cell−cell interactions
DOI: 10.1038/s42003-023-04941-9
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




