Vestimentiferan tubeworms, which live mainly around deep-sea cold seeps and hydrothermal vents, are annelids like earthworms and ragworms. However, instead of consuming food from the outside world, they live through symbiosis with chemosynthetic bacteria in the specialized organ called 'trophosome' and utilize the organic matter produced by these bacteria.
Chemosynthetic bacteria consume hydrogen sulfide (H2S), which is toxic to animals. Therefore, vestimentiferan tubeworms must have the capability to take up hydrogen sulfide and deliver it to the bacteria. Generally, vestimentiferan tubeworms are found at depths of several hundred to several thousand meters, making them difficult to observe and collect. However, Lamellibrachia satsuma, which was discovered in Kagoshima Bay, can be found in shallow waters 100 meters in depth.
The research group led by Associate Professor Chuya Shinzato of the Atmosphere and Ocean Research Institute (AORI), Graduate Student Taiga Uchida of the School of Science at the University of Tokyo, Director Akira Sasaki of Kagoshima City Aquarium, and Curator Ayuta Yamaki of Enoshima Aquarium has sequenced the entire genome of Lamellibrachia satsuma, the world's shallowest-dwelling vestimentiferan tubeworm, and performed gene expression analysis of its various tissues.
The results have revealed that genes involved in biological defense, such as those coding for Toll-like receptors and lysozymes, show characteristic expression patterns in Lamellibrachia satsuma.
Provided by the University of Tokyo
In the present study, comparisons with the genomes of a wide range of other animals showed a variety of characteristics of the vestimentiferan tubeworm genome. For example, the number of genes coding for lysozyme, a protein that attacks bacteria, was higher in Lamellibrachia satsuma than in other annelids.
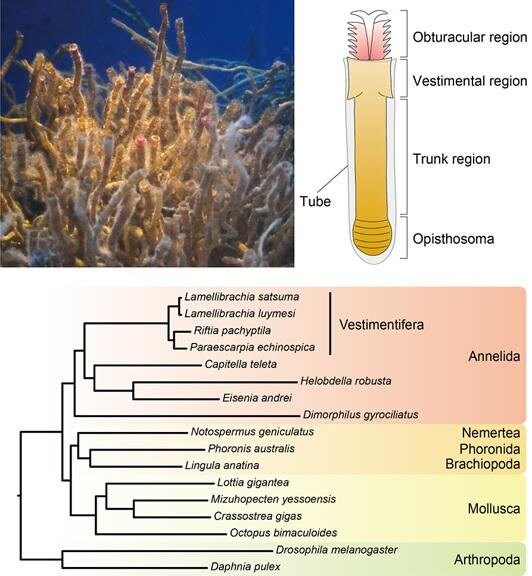
Additionally, a comparison of the gene expression levels in the obturacular region, vestimental region, and trunk region out of four main regions of the vestimentiferan tubeworm body revealed that the majority of lysozyme genes are mainly expressed in the vestimental region. By contrast, the majority of genes coding for Toll-like receptors, which recognize bacteria and viruses and trigger an innate immune response, are primarily expressed in the obturacular region.
The vestimentiferan tubeworm harbors the symbiotic bacteria in the trophosomes present in the trunk region. Such distribution of gene expression suggests that the symbiotic bacteria in the trunk region are spatially isolated from the key factors of biological defense. In other words, it can be speculated that Toll-like receptors and lysozymes are concentrated in the obturacular and adjacent vestimental regions, both of which are most vulnerable to foreign microorganisms, and that they deal with pathogens efficiently while suppressing attacks on bacteria living in the trunk region.
Therefore, which genes are expressed in the trunk region?
Analysis has shown that the number of genes coding for C-type lectins (proteins that bind to carbohydrate chains on the surface of microorganisms) is increased in the genome of the vestimentiferan tubeworm, and that some of these genes are highly expressed in the trunk region. The protein may be involved in coping with foreign microorganisms in the trunk region (where biological defenses are less robust) or in interactions with symbiotic bacteria.
Additionally, genes coding for chitinase (thought to be involved in the decomposition of tubes) and globin (involved in the transport of oxygen and hydrogen sulfide in the body) were also increased in the vestimentiferan tubeworm genome.
The research group has also discovered FIH1-like genes, which are similar to FIH1 genes involved in the regulation of the hypoxic response. The analysis of a wide range of animal species revealed that this FIH1-like gene is unique to the vestimentiferan tubeworm. All of these genes are thought to contribute to the evolution of the vestimentiferan tubeworm's unique lifestyle.
The sequencing of the Lamellibrachia satsuma genome is expected to advance research on the symbiosis between these tubeworms and chemosynthetic bacteria and their adaptation to hydrothermal vents and springs.
Journal Information
Publication: DNA Research
Title: Genomic and transcriptomic analyses illuminate the molecular basis of the unique lifestyle of a tubeworm, Lamellibrachia satsuma
DOI: 10.1093/dnares/dsad014
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




