A research group including Specially Appointed Researcher Shintaro Minami (at the time of research), Specially Appointed Researcher Rie Tatsumi-Koga (Assistant Professor of the Institute for Protein Research (IPR) at Osaka University), and Professor Nobuyasu Koga (Professor of the Institute for Protein Research (IPR) at Osaka University) of the Exploratory Research Center on Life and Living Systems (ExCELLS), in collaboration with RIKEN, Nagoya University, and Osaka University, examined the topology (i.e., the two-dimensional spatial arrangement and loop linking of the α-helices and β-strands) of proteins, and on the basis of both theory and experiments, revealed for the first time that the number of unexplored foldable αβ-type protein topologies is far more than the 400 topologies currently found in nature, perhaps even totaling 10,000. The result was published in Nature Structural and Molecular Biology.
Shintaro Minami, Naohiro Kobayashi, Toshihiko Sugiki, Toshio Nagashima, Toshimichi Fujiwara, Rie Tatsumi-Koga, George Chikenji & Nobuyasu Koga, "Exploration of novel αβ-protein folds through de novo design", Nature Structural & Molecular Biology (2023)
Proteins form specific three-dimensional structures according to their amino acid sequences and exhibit functions based on those structures. The possible number of amino-acid sequence combinations in a protein is vast; for example, a protein consisting of 100 amino acid residues can have approximately 10130 possible sequence combinations. Assuming that there are 10 million kinds of organisms on the Earth and each has 100,000 unique genes, the total number of amino acid sequences would be approximately 1012. This number is much smaller than 10130. It means there is a huge body of proteins in nature that are yet to be explored.
Although more than 10,000 protein structures are experimentally revealed every year, from the viewpoint of protein topology or fold, new topologies are rarely being discovered. Previous studies have theorized whether nature has already exhausted all the possible topologies in which proteins can fold or whether there are still many other folding patterns that nature has yet to exploit. However, these theories have not been experimentally verified.
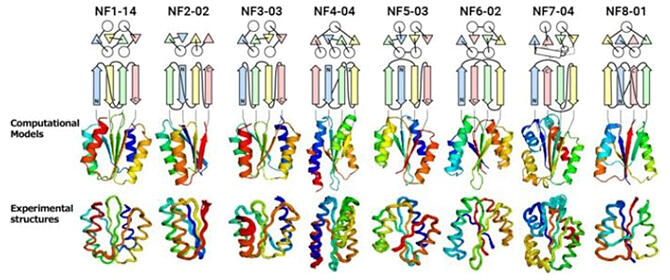
On the basis of physical chemistry and protein structural data, the research team devised rules for theoretically discerning the topologies in which protein molecules can fold. With these rules, they then predicted a total of 12,356 novel foldable αβ-type topologies with β-sheets consisting of 4−8 β-strands, none of which are found in the current data bank of protein structures. Furthermore, using a self-developed computer program with a 'technology that designs protein molecules (including the main chains) from scratch,' the research team designed the protein molecules for 8 novel topologies, including those for proteins consisting of 4-stranded β-sheets that formed a knot. Experimental verification of the folding ability of the designed proteins revealed that all the topological designs could be folded into the structures set out by the computer program. These results indicate that there are approximately 10,000 unexplored foldable αβ-type topologies, suggesting that there could be many foldable topologies existing in nature that are yet to be explored.
The results further suggest that the time period for current biological evolution is too short for nature to explore all possible topologies, and that life on Earth descends from a common ancestor, thus leading to a bias in the emergence of natural protein topologies.
Additionally, by designing proteins that have the approximately 10,000 foldable αβ-type topologies found in this study, it is expected that they will lead to the future development of functional protein molecules that will contribute to medicine and industries.
Journal Information
Publication: Nature Structural & Molecular Biology
Title: Exploration of novel αβ-protein folds through de novo design
DOI: 10.1038/s41594-023-01029-0
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




