A research group led by Professor Katsumi Imada and Graduate Student Shiho Otsubo (both of the Graduate School of Science, Osaka University) and Associate Professor Hiromi Imamura (Graduate School of Biostudies, Kyoto University) have announced that they have succeeded in artificially creating a red fluorescent protein (RFP) by engineering a green fluorescent protein (GFP). They modified the GFP of the stony coral Galaxea fascicularis to RFP and determined the 3D structures. They also identified the amino acids and their conformations important for the formation of the red fluorescent chromophores (color-forming molecules). This achievement is expected to lead to the development of high-performance RFPs suitable for deep biological imaging. The results were published on October 23 in the US journal Proceedings of the National Academy of Sciences.
Provided by Osaka University
Fluorescent proteins are used as biomarkers and biosensors to obtain various information in vivo (i.e., within a living system), such as the expression, distribution, and interaction of specific proteins in living organisms. Until now, the RFPs needed for the long-term observation of thick samples such as tissues and organs were made only from natural RFPs, which are not bright enough. Most natural fluorescent proteins are green, and RFPs have only been found in a few organisms.
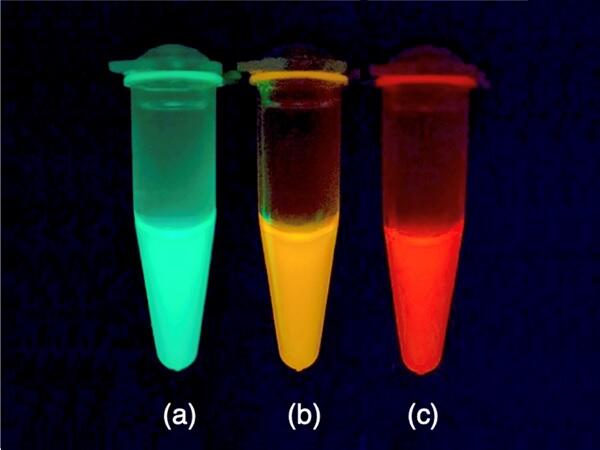
In this study, the research group used a Galaxea fascicularis GFP named AzamiGreen (AG), which has a relatively similar amino acid sequence to that of natural RFPs. They focused on 35 amino acid residues that are highly conserved in natural RFPs but not in GFPs. When these 35 amino acids were changed to those of natural RFPs, the altered AG protein (renamed as AzamiRed0.1 or AR0.1) emitted weak red fluorescence in addition to green fluorescence.
Provided by Osaka University
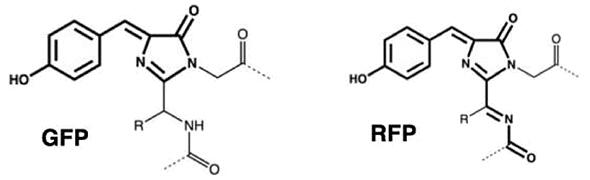
Mutant protein AR1.0, which has one of the highest quantum efficiencies among fluorescent proteins emitting red light (>600 nm), was successfully synthesized by introducing 29 mutations into AG using protein engineering techniques. (Quantum efficiency is the ratio of the number of excitation photons absorbed by a fluorescent material to the number of photons emitted as fluorescence.) Additionally, the research group used X-ray crystallography at SPring-8 (a large synchrotron radiation facility in Harima Science Garden City, Japan) to study the 3D structures of AG and AR1.0 in detail. The structural differences revealed the required arrangement of amino acids inside the fluorescent protein for formation of the red chromophore.
Imamura commented, "At the beginning of this research, we suffered from the fact that red fluorescence did not appear as expected, but we were able to develop the first GFP-derived RFP through continuous investigation. Although the practical aspects of fluorescent proteins tend to be emphasized, the mechanism of chromophore formation is also an interesting research topic. This research should lead to a better understanding of the mechanism of RFP chromophore formation, allowing us to design and develop high-performance RFPs in a logical manner."
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




