A research team led by Associate Professor Shogo Nakano and Graduate Student Hiroki Ozawa from the Graduate Division of Nutritional and Environmental Sciences at the University of Shizuoka, has developed a new tool called "GAOptimizer." This tool enables the computer-based virtual evolution of proteins, leading to the creation of highly functional protein designs. The research team applied the method to three different types of proteins with distinct structures, sequences, and functions and then experimentally confirmed that the artificially designed proteins have superior functions suitable for application compared with the naturally derived proteins used as templates. This tool is anticipated to enhance the design of advanced functionality in natural proteins and expedite the development of protein-based biomaterials for next-generation applications. The study results have been published in Cell Reports Physical Science.
Provided by the University of Shizuoka
The GAOptimizer tool can be downloaded from the website at https://zenodo.org/records/10208126.
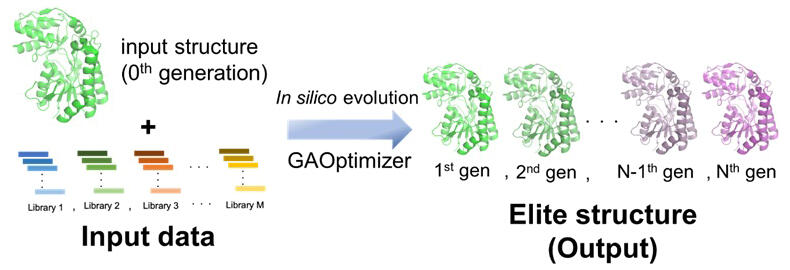
The research team developed GAOptimizer, a new protein design tool that identifies useful mutation points in a target protein to improve its function. The target protein to be improved (template structure) and its homologous sequences (libraries) are used as input data, and the tool then applies genetic algorithms to optimize the combination of multiple useful mutation points to introduce into the target protein.
First, 100 child structures are generated from 30 parent structures by introducing crossover recombination mutations (randomly selecting two structures from the parent structure and crossing them) or random mutations (selecting one structure from the parent structure and introducing a point mutation). Next, a score is calculated for each generated child structure using fitness functions.
One of the fitness functions used is the Rosetta energy unit (REU), which evaluates the stability of the protein structure. The other is the HiSol score, which is independent of structural stability. On the basis of the calculated score values, the child structure with the best fitness is selected as the elite structure and saved as the representative structure of the nth generation. Furthermore, five structures are randomly selected from the 100 generated child structures, and the one with the best fitness among them is used as a candidate for the parent structure.
By repeating this process 30 times, 30 parent structures of the next generation (n+1 generations) are selected from the child structures to complete the evolution of one generation. This calculation is repeated for N generations to complete the virtual evolution.
Through alteration of the fitness function to scores other than REU, the enzyme S-selective hydroxynitrile lyase (S-HNL) underwent virtual evolution. Compared with the natural HNL protein, this modified enzyme demonstrated more than a 10-fold increase in productivity, over a 3-fold increase in catalytic efficiency, and an approximate 5-degree enhancement in heat resistance (Tm). These enhancements indicate that the enzyme has acquired functionalities suitable for applied use.
Journal Information
Publication: Cell Reports Physical Science
Title: Development of evolutionary algorithm-based protein redesign method
DOI: 10.1016/j.xcrp.2023.101758
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




