A collaborative research group, led by Associate Professor Hiroshi Tsugawa of the Department of Biotechnology and Life Science at Tokyo University of Agriculture and Technology, has announced the development of a method to capture lipid diversity in one-third the time required by conventional methods by improving the measurement method and information processing technology of mass spectrometry. The method improves the practical level of "non-targeted lipidomics," which uses mass spectrometry to comprehensively capture lipid components in vivo in a batch. The improved version can be applied to large-scale analyses such as cohort studies. The results were published in the January 11, 2024, issue of Analytical Chemistry, an international academic journal.
Provided by Tokyo University of Agriculture and Technology (Credit: Tokiyoshi Kanako (Fourth-year student, Department of Biotechnology and Life Science, Tokyo University of Agriculture and Technology))
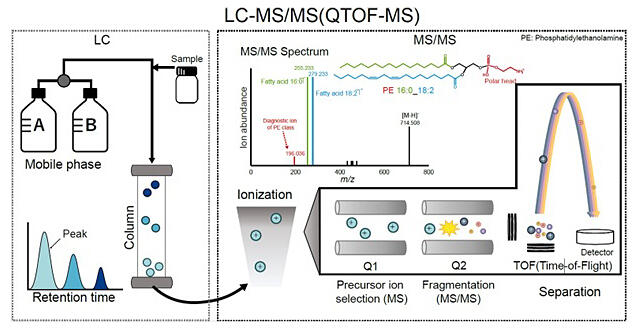
Lipids play a variety of roles in living organisms, not only as a source of energy but also as a component of cell membranes. The diversity and quantitative balance of lipids are important for the development and maintenance of cellular and tissue functions. Lipidomics, a comprehensive analysis of the total lipid population (lipidome) in living organisms, has gained importance for a more detailed understanding of living systems. A technique using liquid chromatography tandem mass spectrometry (LC-MS/MS) is being developed to achieve non-targeted lipidomics that can comprehensively capture lipid diversity without focusing on specific lipid molecules.
LC-MS/MS can unravel the structure of lipids by combining the elution time of compounds using liquid chromatography (LC) with the fragmentation pattern of molecular ions (MS/MS spectra) by mass spectrometry (MS). To increase throughput while maintaining lipid detection sensitivity, the researchers have developed the HybridMS method, which combines two different MS/MS methods: the DDA method, which can obtain highly accurate MS/MS spectra suitable for structural analysis of lipids with the same molecular weight, and the DIA method, which can obtain comprehensive MS/MS spectra regardless of the molecular weight of the lipid molecules.
The DDA method can acquire MS/MS data by prioritizing highly abundant molecules, making it easy to string together relationships and facilitating the structural analysis of new molecules.
While the DIA method can acquire the MS/MS spectra of low-abundance molecules, it is also characterized by the fact that the MS/MS spectra of non-target molecules are also acquired and mixed.
The researchers have also worked on extending the functionality of the analysis program to rapidly capture lipid diversity from the complex datasets generated by the HybridMS method. The developed method was validated using plasma and fecal samples commonly used in clinical research.
As a result, the fast LC + DDA method alone identified 268 lipid molecules in plasma samples and 320 lipid molecules in fecal samples, while the Hybrid MS method identified 468 lipid molecules in plasma samples and 679 lipid molecules in fecal samples, that is, twice as many lipid molecules in the same amount of time. It was also confirmed that there was no loss of quantitation, which had been a problem with the high-speed LC method.
Journal Information
Publication: Analytical Chemistry
Title: Using Data-Dependent and -Independent Hybrid Acquisitions for Fast Liquid Chromatography-Based Untargeted Lipidomics
DOI: 10.1021/acs.analchem.3c04400
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




