The seaweed Hanemo grows to more than 10 cm, despite being a unicellular organism. Because of its extremely high regenerative capacity, it can grow back to its original state even from protoplasm that has diffused into seawater. Although the seaweed has interesting properties, little is known about the mechanisms involved in its regeneration. A group led by Graduate Student Kanta K. Ochiai and Professor Gohta Goshima of the Sugashima Marine Biological Laboratory at the Department of Biological Science of the Graduate School of Science at Nagoya University, in collaboration with the Nagoya University Center for Gene Research, Tokyo Institute of Technology, and the National Institute of Genetics, successfully decoded the genome of the Hanemo with a high degree of accuracy. Not only will research on Hanemo's regeneration mechanisms be advanced, but the development of experimental biology in other large algae can be expected. The results were published in The Plant Journal.
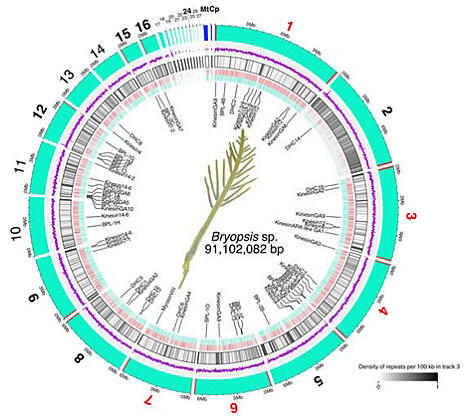
(1) Contigs (cyan) and putative telomeric repeats (red bar, CCCTAAA) are shown. When the repeat was identified in both ends of the contig, the contig number was indicated in red. When just one end had the repeat, the contig was highlighted with a black bold letter. Blue bars indicate organelles of circular genome (mitochondrion: Mt, chloroplast: Cp).
(2) Purple lines indicate G/C content per 10 000 bp. Two grey lines indicate 25% and 75%.
(3) Black bars present non-telomeric repeat sequences.
(4) Red and blue bars indicate genes from Watson and Crick strands, respectively.
(5) Genes analysed in this study.
Provided by Nagoya University
The biggest bottleneck in deciphering the seaweed genome is the difficulty in distinguishing between DNA derived from microorganisms living in symbiosis with the seaweed and DNA derived from the Hanemo. This study separated the two DNA using sterilization operations during the Hanemo culture and precise bioinformatics analysis. As a result, the entire genome of Hanemo was assembled into 29 sequence fragment groups. This is extremely accurate compared to the currently known genomes of large green algae and is a major finding for seaweeds, for which genome information is scarce.
The comparative genomic analysis found 34 kinesin genes and only one myosin gene in Hanemo, although myosin genes are abundant in other large green algae. The administration of drugs inhibited the kinesin-microtubule system and suppressed chloroplast movement in Hanemo cells, suggesting that the kinesin-microtubule system leads to Hanemo's intracellular trafficking. In addition, 15 lectin genes, BPL-1, which is believed to be involved in the aggregation of Hanemo protoplasm during regeneration, were found. No BPL-1 genes were found in other large green algae with low regenerative capacity, suggesting the possibility that the large duplication of the BPL-1 gene maintains Hanemo's high regenerative capacity. Green algae, including Hanemo, come from the same ancestors as plants.
This study is particularly important in examining how the mechanisms involved in the high regenerative capacity and transport of materials in large green algae, including Hanemo, differ from those in plants. Future research is expected to lead to new developments in algae science, which has lagged behind research compared to plant and animal research.
Journal Information
Publication: The Plant Journal
Title: Genome sequence and cell biological toolbox of the highly regenerative, coenocytic green feather alga Bryopsis
DOI: 10.1111/tpj.16764
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




