Using cryo-electron microscopy, Graduate Students Yutaro Shuto, Ryoya Nakagawa (at the time of research) and Mizuki Hoki (at the time of research), and Professor Osamu Nureki of the Graduate School of Science at the University of Tokyo, have determined the three-dimensional structures of a "prime editor," a genome editing tool combining the CRISPR-Cas enzyme and reverse transcriptase, during the process of reverse transcription of a DNA target. The obtained structures and biochemical analyses revealed, for the first time in the world, the details of the sequential processes by the prime editor from the initiation of reverse transcription to the elongation of the DNA strand and termination of reverse transcription. The results were published in Nature.
Provided by the University of Tokyo
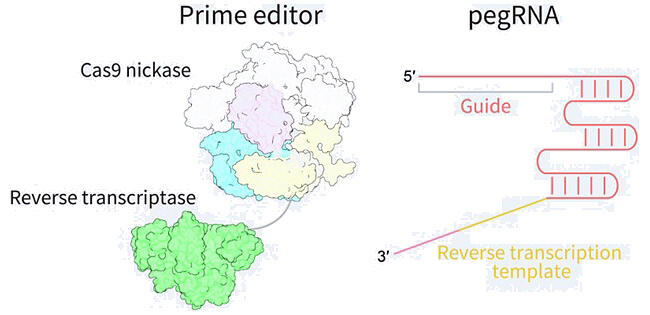
As a genome editing technology that artificially modifies genomic information, CRISPR-Cas9 has been widely used to create genetically modified organisms for research, crop variety improvement, gene therapy, and other purposes. However, the accuracy of editing and the safety of cutting double-stranded genomic DNA are issues in gene therapy. Prime editing, which was reported in 2019, is a new Cas9-based genome editing technology that does not "cut" double strands. In this approach, a prime editor consisting of a Cas9, which does not cut one of the two strands, and reverse transcriptase, forms a complex with a modified guide RNA (pegRNA) to which the sequence to be edited is added as a template sequence. In turn, the prime editor induces reverse transcription of the template sequence at the targeted position to replace the genomic information accurately. In this manner, the prime editor functions like a word processor. However, the detailed molecular basis of how prime editors perform pegRNA-dependent reverse transcription was unclear.
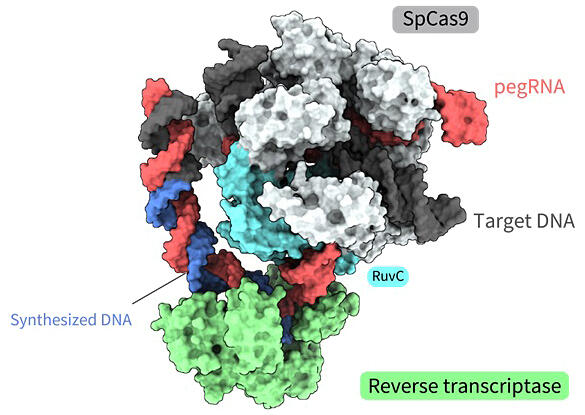
Using cryo-electron microscopy, the research group determined multiple three-dimensional structures of a prime editor involved in pegRNA-dependent reverse transcription of target DNA. The obtained structures revealed a process in which reverse transcriptase binds to the RNA-DNA heteroduplex formed along the RuvC domain of Cas9 and performs reverse transcription with almost no change in position relative to Cas9. The obtained three-dimensional structures and biochemical analyses suggested that the reverse transcriptase performs extra reverse transcription reactions beyond the expected position and terminates reverse transcription upon collision with Cas9. Furthermore, since such extra reverse transcription leads to the insertion of unwanted sequences through prime editing, they successfully developed a modified pegRNA to prevent unwanted insertion by rational modification based on the obtained three-dimensional structures.
Provided by the University of Tokyo
The findings are expected to contribute significantly to further understanding of the principles of prime editing and the design and development of next-generation prime editors with even higher levels of accuracy and editing efficiency.
Journal Information
Publication: Nature
Title: Structural basis for pegRNA-guided reverse transcription by a prime editor
DOI: 10.1038/s41586-024-07497-810.1
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




