An international collaborative research group led by RIKEN and other institutions has succeeded in analyzing gene promoters and enhancers of diverse human helper T cells at the single-cell level and systematically clarified how these cells are involved in the development of various immune diseases. In addition to the enhancers and target disease genes clarified in this study, the code for the single-cell enhancer analysis method (ReapTEC approach) has been published through DDBJ. Team Leader Yasuhiro Murakawa said, "We hope that this method will be used, because enhancer RNAs can be identified by reanalyzing previously obtained gene expression/analysis data." The findings are expected to contribute to the development of new treatments for autoimmune and allergic diseases. The work was published in Science.
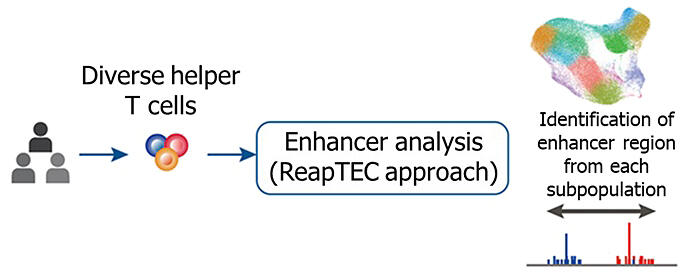
A unique methodology (ReapTEC approach) was developed to identify RNA transcription start sites at the single-cell level and simultaneously analyze promoter and enhancer activity and was then applied to approximately one million helper T cells in humans to generate an enhancer activity map of helper T cells.
Provided by RIKEN
An enhancer is a gene expression switch that activates a target gene. Genetic mutations are enriched in enhancers, and these mutations are involved in the development of diseases by altering the quantity, rather than the quality, of genes. An international collaborative research group led by: Research Associate Akiko Oguchi (Specially Appointed Research Scholar, ASHBi, Kyoto University), Visiting Researcher Shuichiro Komatsu, and Team Leader Yasuhiro Murakawa (Professor, ASHBi, Kyoto University) of the RIKEN-IFOM Joint Laboratory for Cancer Genomics, RIKEN Center for Integrative Medical Sciences; Deputy Team Leader Akari Suzuki and Team Leader Kazuhiko Yamamoto (Director of the RIKEN Center for Integrative Medical Sciences) of the Laboratory for Autoimmune Diseases, RIKEN Center for Integrative Medical Sciences; and Team Leader Chikashi Terao of the Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences has developed a new method for identification of the RNA transcription initiation sites at the single-cell level and simultaneous activity analysis of enhancers and gene promoters using a modified information processing technique to accurately capture signals derived from the cap structure added to the first nucleotide of RNA. By applying this method to approximately 1 million helper T cells collected from humans, the research team identified RNA transcription initiation sites, which are dynamically changing, in individual helper T cells and measured RNA expression levels.
The team found rare subpopulations that have not been previously reported and revealed the diversity of helper T cells. Moreover, they comprehensively identified the gene promoters and enhancers that were activated in each helper T cell subpopulation. They then identified a total of more than 60,000 enhancers that were activated in helper T cells and created an enhancer activation map for each helper T cell subpopulation.
Large-scale genome-wide association studies (GWAS) have identified hundreds of genetic mutations associated with various autoimmune and allergic diseases. Their analysis involved superimposition of the sites of these genetic mutations associated with the immune diseases on the enhancer regions activated in helper T cells. The results showed that genetic mutations associated with autoimmune and allergic diseases such as multiple sclerosis, systemic lupus erythematosus, rheumatoid arthritis, asthma and inflammatory bowel disease were highly enriched in the enhancer regions of helper T cells. They also identified approximately 600 helper T cell enhancers (disease enhancers) with genetic mutations associated with immune diseases.
An enhancer works after it comes spatially close to its target gene. Therefore, the team generated Micro-C data with the world's highest level of depth of 40 billion reads. The Micro-C method can determine the three-dimensional structure of the genome with a high base resolution at the nucleosome level. The data were used to comprehensively deduce the target genes of disease enhancers. They deduced the target genes of respective disease enhancers, systematically analyzed the molecular pathways leading to the onset of autoimmune and allergic diseases and identified many new disease-associated molecules. For example, activation of an enhancer associated with inflammatory bowel disease resulted in specific induction of the IL7R gene expression. In fact, blockers of the IL7 receptor have shown therapeutic efficacy against ulcerative colitis and are tested in phase II clinical trials. While drug targets are said to be exhausted, the identification of a new group of immune disease-related molecules through this approach raises expectations for applications such as new molecularly targeted drugs, cell therapies and personalized medicine.
In this study, the research group identified associations of helper T cells with autoimmune and allergic diseases. This approach is also applicable to other diseases, such as lifestyle-related diseases, cardiovascular diseases and psychiatric disorders. They also analyzed killer T cells and B cells as well as enhancer regions related to brain diseases.
Journal Information
Publication: Science
Title: An atlas of transcribed enhancers across helper T cell diversity for decoding human diseases
DOI: 10.1126/science.add8394
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




