Research Scientist Saori Takahashi and Team Leader Ichiro Hiratani of the Laboratory for Developmental Epigenetics at the RIKEN Center for Biosystems Dynamics Research, in collaboration with Assistant Professor Hirohisa Kyogoku of the Graduate School of Agricultural Science at Kobe University (also a visiting scientist at the RIKEN), and Professor Shin-ichiro Takebayashi of the Graduate School of Bioresources at Mie University, discovered a specialized genomic DNA replication mode in early mouse embryos immediately after fertilization. A unique single-cell whole-genome analysis technique was used to analyze DNA replication in early mouse embryos.
The study revealed that in embryos in the 1- and 2-cell post-fertilization stages, the DNA replication mode differed from that in somatic cells, and DNA strands were synthesized slowly, whereas 4-cell embryos had an intermediate unstable replication mode and a high frequency of chromosome segregation errors. These findings are expected to contribute to understanding why chromosome segregation errors occur more commonly during early development in mammals and establish a foundation for reproductive medicine. The results were published in the international journal Nature on August 28.

Provided by RIKEN
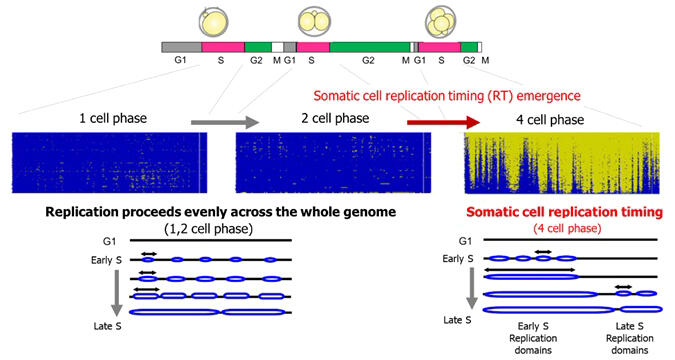
In multicellular organisms, a single fertilized egg undergoes development through repeated cell division cycles during which genomic DNA is replicated and segregated into two daughter cells before cell division. Recent studies in somatic cells have shown that genomic DNA replication in somatic cells is strictly regulated in terms of replication timing, such as which regions are replicated and when they are replicated. The regions replicated in the first and second halves of the S phase of the cell cycle are clearly distinct.
Meanwhile, the way in which DNA replication is regulated during early embryonic development is known and it is also known that the frequency of chromosome segregation errors during early development in mammals is higher than that in somatic cells. However, the underlying mechanisms remain unknown because single-cell analysis is difficult.
Previously, the research group developed a single-cell whole-genome DNA replication sequencing technology (scRepli-seq) in 2019. This technology allows genome-wide analysis of the genomic DNA doubling process in a single proliferating cell.
In this study, this method was applied to analyze early mouse embryos immediately after fertilization from the 1-cell stage to the blastocyst stage. The results illustrate that at the 1- and 2-cell stages, the replication timing that can be seen in somatic cells was absent, and the entire genome was replicated uniformly. The replication timing regulation was noted in the 4-cell stage for the first time, and replication timing was maintained after that. Moreover, the distribution of replication initiation points and replication fork speeds (the rate at which DNA strand is synthesized) in early embryos were examined using the DNA fiber method.
In the 1- and 2-cell stages, the replication initiation points were distributed at 1 per 15,000 to 20,000 base pairs, and the rate of replication was approximately 30 base pairs per minute. These values are more than five times denser and more than 30 times slower than respective values in somatic cells. In contrast, the 8-cell embryos replicated at the rate of 760 base pairs per minute, which is similar to that of somatic cells.
In the 4-cell stage in between, the regulation of replication timing is similar to that found in somatic cells. However, this rate was similar to that in the 1- and 2-cell stages, indicating that the 4-cell stage is a transitional replication stage. scRepli-seq was used to verify whether the frequency of chromosome segregation errors during early development was higher compared to that in somatic cells.
The results showed that the frequency of chromosome segregation errors during the 4-to-8-cell division was approximately 13%, which is a high value unlike somatic cells. The error rates in the 1- and 2-cell stages and the 8- to 16-cell stages were both a few percent. They also confirmed that most of the chromosome segregation errors were structural abnormalities caused by chromosome breaks. These broken chromosomal DNA regions were concentrated in regions replicated in the second half of S phase.
The results suggest the possibility that unreplicated regions are left as-is during chromosome segregation in the 4-to-8-cell division, resulting in segregation errors. Live cell imaging of the DNA replication process revealed that the S phase was longer in 4-cell embryos and even longer in 4-cell embryos with chromosome segregation errors. Therefore, nucleotide precursors, which are used for DNA synthesis, were added to the culture medium to increase the speed of DNA replication at replication fork. As a result, high frequency of chromosome segregation errors during the 4-to-8-cell division stage was reduced to a few percent. Specialized transitional DNA replication at the 4-cell stage was proven to be responsible for noticeable chromosome segregation errors during early development in mammals.
Provided by RIKEN
Kyogoku said, "Our research is still at the starting point. We now know that the replication mode is very specialized, but there are many more questions, such as what mechanisms underlie this specialized mode of replication and whether these phenomena are evolutionarily conserved in other organisms, including humans. Furthermore, the embryos properly develop into offspring despite the high frequency of chromosome segregation errors, suggesting the presence of certain mechanisms to restore these errors. I see this discovery as a starting point for new research. I believe that this has been a very interesting study, including its development into reproductive medicine research."
Journal Information
Publication: Nature
Title: Embryonic genome instability upon DNA replication timing program emergence
DOI: 10.1038/s41586-024-07841-y
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




