A research group led by Professor Seiji Kojima, Professor Takayuki Uchihashi, and Professor Emeritus Michio Homma of the Graduate School of Science at Nagoya University, in collaboration with Assistant Professor Norihiro Takekawa of the Graduate School of Science at Osaka University, and the Institute for Protein Research and the Graduate School of Frontier Biosciences at Osaka University, has announced that they solved the structure of the complex serving as the scaffold for formation of the bacterial motor organ "flagellum" (S ring). Detailed information on the bacterial flagellar structure is expected to contribute to preventing and treating infectious diseases and developing biological nanomachines. The results were published in the international journal mBio on September 6.
Provided by Dr. Takekawa Norihiro, Osaka University
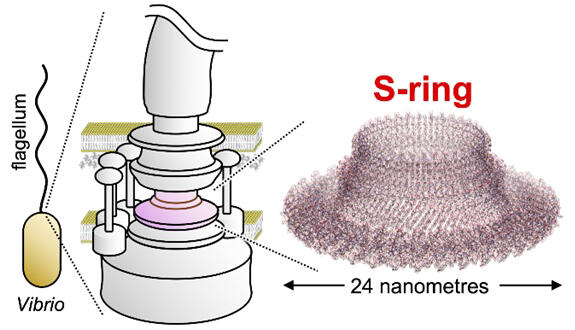
The flagellum serves as a device for bacterial movement, and its rotation allows the bacterium to swim in the fluid. Flagella are found in many bacterial species. The flagellum has a complex structure consisting of 20,000-30,000 protein molecules. A flagellar motor at its base acts as a rotary motor and consists of two structures, M and S rings. These rings are made from dozens of FliF (protein molecules) during the earliest stages of flagellar formation and have been shown to act as scaffolds for flagellar formation and directly affect motility.
Until now, the detailed structure of the flagellar S-ring has only been solved in Salmonella spp. This will provide important insights into the evolution of mechanisms underlying bacterial motility. In this study, the structure of the flagellar S-ring of the bacteria was solved in great detail using cryo-electron microscopy. The results showed that both Vibrio and Salmonella S rings comprised 34 FliF molecules and had the "RBM3 domain" and "β-collar" as basic components.
Meanwhile, the interaction between the "RBM3 domains" in the Vibrio S-ring is weaker than that in the Salmonella S-ring, reflecting the Vibrio-specific inefficient flagellar formation in previous studies. The tilt angle at the base of the "RBM3 domain" or "β collar" was different between the Vibrio and Salmonella S-rings. However, due to the difference in the shape of the "β2-β3 loop," both had near perpendicular angles at the tip of the "β collar." A detailed understanding of the structure and function of bacterial flagella could lead to the development of techniques to inhibit or, conversely, promote bacterial motility.
Kojima said, "The preparation of the samples we observed in this study was a long-standing challenge, but we overcame this difficulty. We could finally succeed in our research by combining all team members' strengths. Moving forward, we build on these results and further basic research toward controlling bacterial motility and pathogenicity. Ultimately, we hope to contribute to developing new therapeutics for infectious diseases and nanomachines."
Journal Information
Publication: mBio
Title: Structural analysis of S-ring composed of FliFG fusion proteins in marine Vibrio polar flagellar motor
DOI: 10.1128/mbio.01261-24
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




