The research method used to comprehensively examine the locations, types, and amounts of genes expressed in tissue samples is called "spatial transcriptomics (ST)." Research using ST has been progressing in recent times. The advent of new analytical techniques in recent years has made it possible to understand, at the molecular level, cellular functions in tissues and the relationship between changes in those functions and diseases. However, ST had problems, such as very high costs for experiments and the data complexity, which precluded analysis by non-experts.
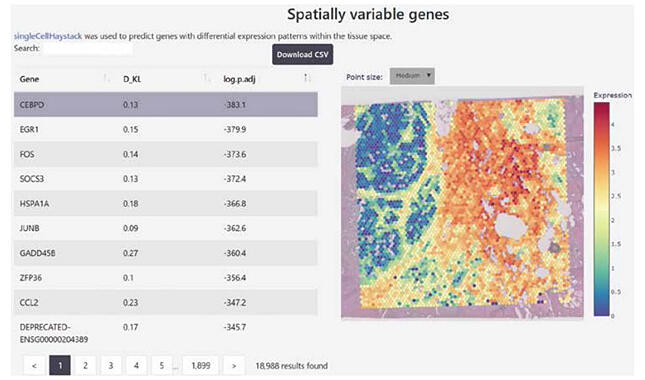
Associate Professor Alexis Vandenbon of the Institute for Life and Medical Sciences at Kyoto University has developed and released "DeepSpaceDB." This database (DB) complies a collection of ST data from various tissues that have been published to date. Using this DB, researchers can easily view and analyze the ST data without performing the experiments themselves. As of September 2024, the DB contains data on 626 human and 412 mouse samples and covers almost all the data obtained on "10x Visium," a major ST platform published by the U.S. National Center for Biotechnology Information (NCBI) and others.
DeepSpaceDB is built in such a manner to facilitate careful examination and analysis of sampled tissues, disease states, and other information. Users can examine the expression sites of any gene or the sites where a certain function is activated or inactivated. This DB is expected to contribute to elucidation of the etiology and pathophysiology of diseases, because it can be used to know, for example, where in cancer tissue one can find cells of what state.
DeepSpaceDB link: https://genomics.virus.kyoto-u.ac.jp/deepspacedb/
Display of the spatial expression distribution of arbitrary genes