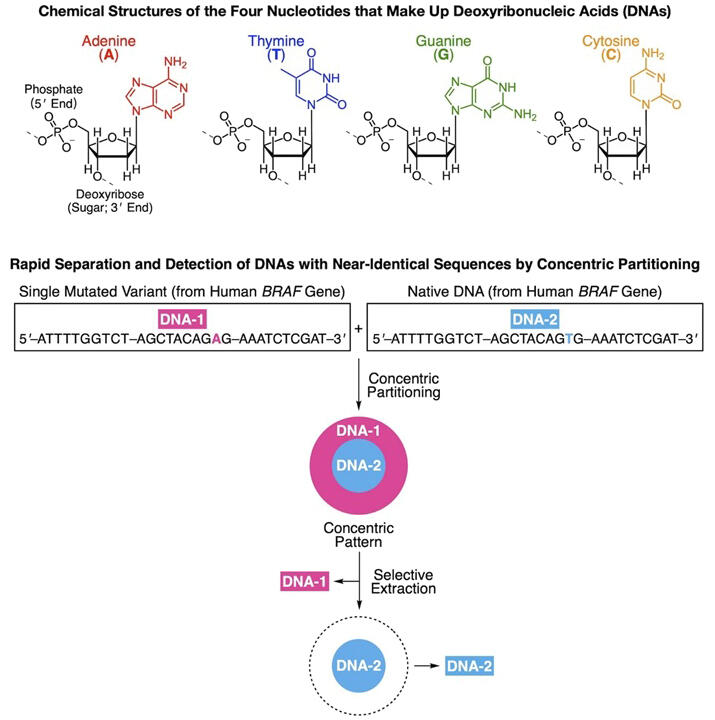
DNA and RNA carry the basic genetic information required for life in organisms. For example, the substitution of just a single base out of approximately 2300 bases in the human BRAF gene can lead to the development of multiple cancers. Therefore, techniques to separate nucleic acids with similar structures are important but difficult to develop. A research group led by Distinguished University Professor Takuzo Aida at the Tokyo College of the University of Tokyo Institutes for Advanced Studies and Project Researcher Hao Gong at the Graduate School of Engineering at the University of Tokyo has discovered that nucleic acids with similar structures can separate in concentric circles on a glass plate using only salt water. Furthermore, they have succeeded in isolating these nucleic acids easily. The study was published in Nature.
Provided by the University of Tokyo
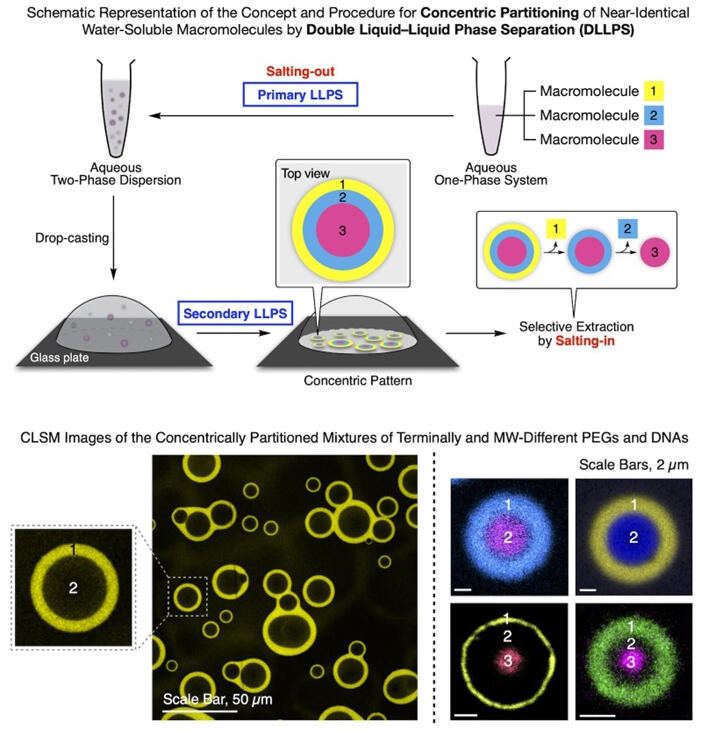
The salting-out effect, in which a solute is separated from an aqueous solution by adding salt, has been used in industry for more than 200 years. When the research group did a completely different experiment using polyethylene glycol (PEG) with a fluorescent molecule attached to its termini, a solution containing ammonium sulfate accidentally fell onto a glass plate. Numerous doughnut-shaped fluorescent circles were formed on the glass plate.
On closer examination, the donut's center was packed with PEG with no dye at the termini. Furthermore, when a mixture of three different polymers was treated similarly, the three polymers were separated into three concentric circles. They found that the mechanism of this phenomenon was explainable as follows. Adding ammonium sulfate to an aqueous solution of a polymer mixture causes liquid-liquid phase separation (LLPS), leading to the dispersion of the polymers in the aqueous solution. When it is dropped onto the glass plate, a second LLPS occurs, and a film of ammonium sulfate is formed first on the glass plate. Then, the polymers form concentric circles in descending order of affinity with it, and finally, the polymer with the lowest affinity is left at the center.
Aida said, "I thought that this phenomenon could be used in principle to separate nucleic acids, so I tried it and found that even nucleic acids with only a single nucleotide difference can be phase-separated. We also found that since LLPS relies on the salting-out effect, the nucleic acids could be extracted in descending order simply by rinsing the plate with water containing different salt concentrations."
In this study, they tested if a single base mutation in 30- and 50-base sequences derived from the human BRAF gene was enough for separation and succeeded in isolating the nucleic acids at 75% purity in one operation (about 10 minutes by hand) and 97% purity in three operations. Automating this technique, which allows for rapid and low-cost detection and separation of mutants, would contribute to major advances in life science.
"We are considering collaboration with analytical manufacturers and other companies," Aida added.
Provided by the University of Tokyo
Journal Information
Publication: Nature
Title: Near-identical macromolecules spontaneously partition into concentric circles
DOI: 10.1038/s41586-024-08203-4
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




