A research group led by Assistant Professor Satoshi Ninagawa and Graduate Student Masaki Matsuo of the Biosignal Research Center at Kobe University, Professor Kazutoshi Mori (at the time of the study) and Graduate Student Deng Ying (at the time of the study) of the Graduate School of Science at Kyoto University, Professor Koichi Kato of the Exploratory Research Center on Life and Living Systems at the National Institutes of Natural Sciences, and Associate Professor Hirokazu Yagi of the Graduate School of Pharmaceutical Sciences at Nagoya City University, has announced their research results revealing that the fate of proteins in the endoplasmic reticulum is determined by a "tug-of-war" between structure-forming and degradation factors via glycans. This discovery was made by studying the function of "UGGT1," an endoplasmic reticulum glycosyltransferase that promotes protein structure formation. The findings are expected to be useful for proposing new therapeutic and prophylactic approaches to human diseases. The results were published in the international journal eLife on December 10.
Provided by Kobe University
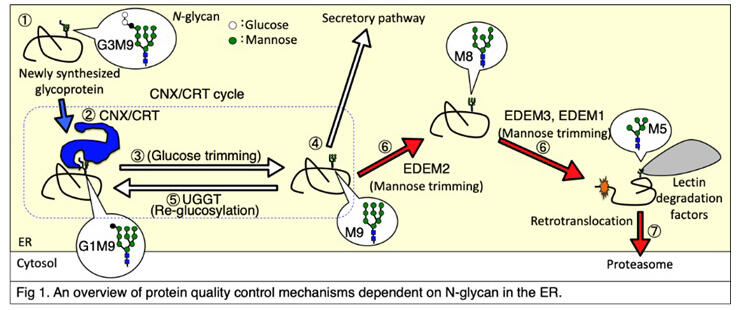
In the endoplasmic reticulum, one of the cellular organelles, one-third of all proteins, including a group of secretory pathway proteins comprising membrane proteins and secretory proteins, are biosynthesized. N-glycans are added to about 80% of them, and strict quality control is performed via these glycans. The pathways of protein structure formation and degradation in the endoplasmic reticulum have been shown to differ greatly depending on the presence or absence of N-glycans.
N-glycans comprise three glucose and nine mannoses (G3M9) and are added to proteins. If the N-glycans of proteins are processed to GM9, a lectin chaperone (CNX) or CRT recognizes their structures to facilitate the protein structure formation. If proteins with M9 have the proper protein structure, they move on to the next step of the secretory pathway. If the proteins with M9 do not have the proper protein structure, glucose is added to M9 by the glucosyltransferase UGGT1 to form the proteins with GM9, which undergo the same cycle again.
On the other hand, if the proteins with M9 do not establish the proper protein structure after the repeated cycles, mannose residues are removed from the N-glycan by EDEM2, and the proteins are finally recognized by lectin degradation factors for degradation. Meanwhile, it was unclear how the fate of these proteins was determined.
It has been proposed, but not fully proven, that UGGT1, which is involved in the structure formation of the endoplasmic reticulum, contributes to protein structure formation but does not affect the timing of degradation. To verify this, the research group generated UGGT1 gene-disrupted cells (KO cells) and examined them.
The results showed that the membrane protein ATF6, which was used as a degradation substrate, did not undergo the N-glycan reglucosylation and degraded at a markedly accelerated rate in UGGT1-KO compared to WT cells. The glucosylation enzyme UGGT1 was found to decelerate degradation. Furthermore, while UGGT1 had no major effect on proteins with stable structures and long half-lives, UGGT1 suppressed the degradation of proteins with incomplete or unstable structures. Previous study findings were summarized, indicating that the quantitative balance of structure-forming and degradation factors determines whether structure formation or degradation is more prone to occur.
The tug-of-war between structure-forming and degradation factors via glycans was found to determine the fate of endoplasmic reticulum proteins. As endoplasmic reticulum function has been implicated in >60 human diseases, including Alzheimer's disease, cancer, and diabetes mellitus, the findings are expected to contribute to the development of new therapeutic and other means for these diseases.
Ninagawa said, "I am pleased to understand the very important mechanism of determining the fate of biosynthesized proteins in the endoplasmic reticulum, that is, whether they undergo structure formation or degradation. We were very surprised when we found that UGGT1, which had previously been thought to have absolutely no role in degradation, noticeably suppressed the degradation of ATF6 and also suppressed the degradation of other structurally unstable substrates."
Journal Information
Publication: eLife
Title: UGGT1-mediated reglucosylation of N-glycan competes with ER-associated degradation of unstable and misfolded glycoproteins
DOI: 10.7554/eLife.93117.4
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




