A research group led by Assistant Professor Takanori Harashima, Assistant Professor Akihiro Otomo, and Professor Ryota Iino of the Institute for Molecular Science has announced that they successfully improved the motion performance of DNA-nanoparticle motors, which represent a type of artificial molecular motors, based on the analysis of elementary processes of motion and chemical reactions. Their artificial molecular motors simultaneously accomplished a motion speed of 30 nm/second and a run-length of 3 µm, which are comparable to the motion performance of naturally occurring motor proteins. The findings are expected to contribute to the development of molecular computers that perform calculations based on the movement of artificial molecular motors and testing systems that detect infection- and disease-related molecules with high sensitivity using the movement of artificial molecular motors. The results were published online in the international journal Nature Communications on January 16.
Provided by the Institute for Molecular Science
Kinesin and myosin are known as naturally occurring motor proteins. They play important roles in intracellular substance transport, muscle contraction, and other biological processes through high-speed movements at 10-1000 nm/second using ATP (adenosine triphosphate) molecules as energy sources. Studies are being conducted to design artificial molecular motors based on these motor proteins and apply them to nanoscale transport devices and other molecular machines. In particular, there are high expectations for the development of DNA-nanoparticle artificial motors as they offer high flexibility in designing base sequences and structures.
Meanwhile, DNA-based artificial molecular motors reported previously had a slow-motion speed of 0.1 nm/second. Although artificial molecular motors based on DNA modified with gold nanoparticles have achieved the fastest motion speed of approximately 1 nm per second, further speed improvements are still needed.
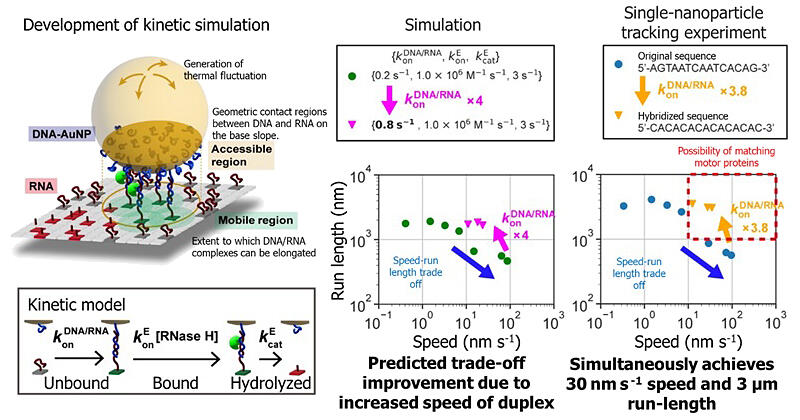
In this study, the research group aimed to identify and improve the bottleneck process of the movement of DNA-nanoparticle (comprising isotropic spherical gold nanoparticles) motors. High-speed, high-precision single-particle tracking revealed that DNA-nanoparticle motors do not move continuously at a constant speed but instead exhibit pause-step motion similar to motor proteins, in which they alternately repeat stopping (pauses) and instantaneous movements (steps). They also found that the slow-motion speed was because the pause length of 70 seconds was longer than that of motor proteins. Elementary processes of chemical reaction proceed while the motors were paused. Among the chemical reaction processes, RNaseH binding was identified as the bottleneck process. When this binding process was accelerated, the pause length shortened to approximately 0.2 seconds; however, the run-length decreased due to a trade-off mechanism. To identify the cause, a simulation was developed to reproduce the movement.
The DNA/RNA sequence was designed based on the prediction that a fourfold increase in the rate of DNA/RNA duplex formation (hybridization) would allow for increasing both motion speed and run-length without a trade-off. As a result, the rate of duplex formation was improved by 3.8-fold.
Experiments with a motor designed using this sequence demonstrated a motion speed of 30 nm/second and a run-length of 3 µm, matching those of natural motor proteins and setting a record for nanosized artificial molecular motors.
Harashima said, "DNA is not only a biomolecule that controls genetic information but also a promising component of nanomaterials such as artificial molecular motors. For example, we believe the findings of this study can be applied to developing transport systems capable of precisely moving objects at the nanoscale and molecular computing that uses motion for performing calculations."
Journal Information
Publication: Nature Communications
Title: Rational engineering of DNA-nanoparticle motor with high speed and processivity comparable to motor proteins
DOI: 10.1038/s41467-025-56036-0
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




