A joint research team from NTT and Associate Professor Tomohiro Shimada of Meiji University's School of Agriculture has identified, for the first time in the world, multiple genes that contribute to the long-term survival of microorganisms in soil, using Escherichia coli as a model. These genes are gaining attention for their potential to serve as a foundational technology for reducing environmental impact, including greenhouse gas emissions from soil. The study was published in Scientific Reports.
Provided by NTT and Meiji University
CO2 emissions from the land, including soil, are about 12 times higher than emissions from human activities. Nitrous oxide (N2O), which has a greenhouse effect that is approximately 290 times greater than that of CO2, is produced by excessive addition of chemical fertilizers to the soil and the activity of soil microorganisms. There is a need to develop technologies to appropriately control microbial activity in soil to reduce environmental burdens. However, conventional methods that change physical properties, such as soil hardness, water retention, and air permeability, and chemical properties, such as soil pH and the type and input of nutrients, can only change the overall amount of the diverse microbial flora in the soil but cannot alter the amount of specific microbial species. Consequently, it has been difficult to effectively reduce greenhouse gas emissions from the land.
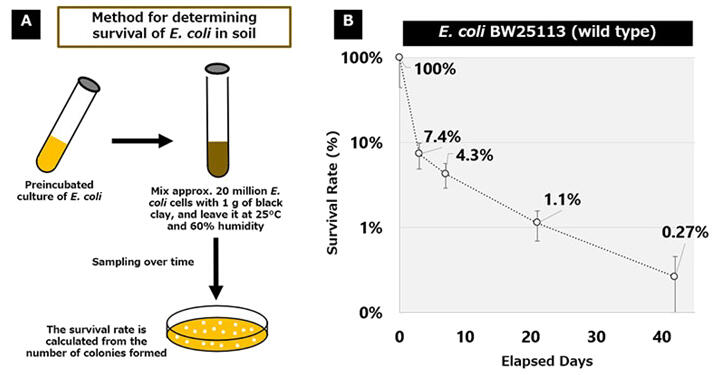
To address this challenge, the joint research group focused on identifying genes that determine the survivability of microorganisms in soil as the first step toward developing technologies to individually control the survivability of specific microorganisms. In this study, they identified such genes using E. coli. First, they established an assay to evaluate the long-term survivability of E. coli in soil. Approximately 20 million E. coli cells mixed with 1 gram of soil were placed in a chamber set 25℃ and 60% humidity. Since the conventional technique using polymerase chain reaction (PCR) cannot distinguish between living and dead cells, truly viable cells were counted as the number of colonies formed by living cells on an agar plate at 0, 3, 7, 21, and 42 days after the incubation. The results showed that the survival rates relative to day 0 (= 100%) decreased to 7.4% (3 days), 4.3% (7 days), 1.1% (21 days), and 0.27% (42 days), respectively.
Using this method, they attempted to identify genes involved in the survivability of E. coli in soil. By focusing on approximately 300 transcription factors possessed by E. coli, the relationship between these transcription factors and survivability in soil was examined using E. coli strains lacking the respective transcription factor genes. By comparing their survivability with that of the wild-type strain, it can be inferred that each transcription factor plays a negative or positive role in the survival based on whether the survivability of the deficient strain was higher or lower. After analyzing 294 deficient strains, they identified 14 strains in which the loss of a specific transcription factor resulted in altered survivability, with 4 showing increased survivability and 10 showing decreased survivability. Among these genes, previous studies have identified the rpoS gene as a transcriptional regulator that affects long-term survivability in soil. Therefore, the involvement of the remaining 13 genes in long-term survivability in soil was demonstrated for the first time in this study. Furthermore, based on the knowledge of the gene functions accumulated in previous studies, they summarized the functions of the 14 identified genes in microorganisms. As a result, these genes could be classified into those involved in stationary phase stress (Dps and RpoS), nitrogen metabolism (Lrp and RpoN), carbon metabolism (CRP and Mlc), osmotic stress (NhaR and OmpR), and other specific functions (AbgR, CadC, Cbl, DinJ, lhfA, and Ygbl).
These findings revealed that microorganisms utilize gene groups related to stationary-phase adaptation, osmotic stress response, and the metabolism of carbon and nitrogen sources to control their survivability in the soil. Additionally, these transcription factors are highly conserved across microbial species, indicating that their functions are fundamental to microorganisms. The results of this study are expected to be applicable not only to E. coli but also to other microorganisms responsible for nutrient cycling in soil. This could contribute to reductions in greenhouse gas emissions, excess nitrogen release, and the reliance on chemical fertilizers.
Journal Information
Publication: Scientific Reports
Title: Identification of a comprehensive set of transcriptional regulators involved in the long-term survivability of Escherichia coli in soil
DOI: 10.1038/s41598-025-85609-8
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




