Biomolecules such as proteins and nucleic acids form biomolecular condensates through phase separation phenomena in cells. Biomolecular condensates play important roles in cells, such as ribosome assembly and RNA protection and degradation. Furthermore, the molecular interactions important for condensate formation are closely related to neurodegenerative diseases such as Alzheimer's disease. Understanding these interactions is essential for elucidating the pathogenesis of and developing therapeutic means for these diseases. Molecular dynamics simulations, particularly coarse-grained models, are suitable for research on biomolecular condensates because they can simultaneously handle a large number of biomolecules. Therefore, high-accuracy coarse-grained models for proteins have been proposed. However, proteins and RNA in cells often form biomolecular condensates. There is a need to develop RNA models for research into protein behavior.
A research group led by Graduate Student Ikki Yasuda of the Graduate School of Science and Technology, Associate Professor Eiji Yamamoto and Professor Kenji Yasuoka of the Faculty of Science and Technology at Keio University, and Professor Kresten Lindorff-Larsen at the Department of Biology at the University of Copenhagen, Denmark, has developed a coarse-grained molecular model of RNA and succeeded in simulating biomolecular condensates formed in protein-RNA mixtures. The model allowed for the elucidation of a new phase-separation mechanism involving RNA at the molecular level. The study was published in the Journal of Chemical Theory and Computation.
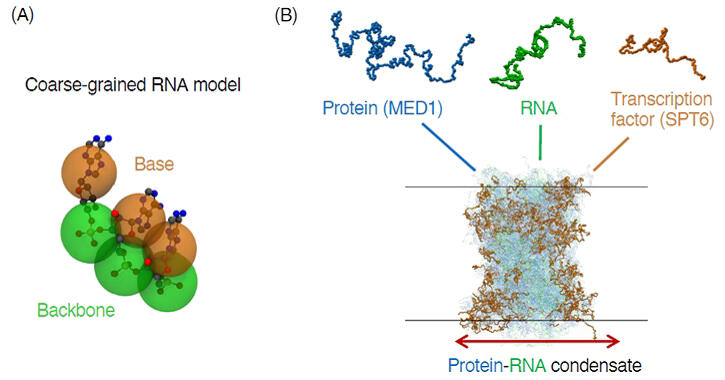
(B) Molecular dynamics simulation of transcriptional condensate formed by proteins and RNA.
Provided by Keio University
In this study, they developed a coarse-grained model of structurally disordered, flexible RNA to represent its physicochemical properties. The model was adjusted to properly reproduce local RNA geometry and intramolecular interactions based on experimental data and all-atom simulations.
As a result, they succeeded in reproducing the formation of biomolecular condensates in simulations involving a large number of proteins and RNAs. Furthermore, focusing on transcriptional condensates, they simulated the formation of mixed condensates consisting of the intrinsically disordered region of a mediator (MED1) and RNAs and compared the simulation results with experimental results.
The simulations reproduced the experimental results showing that the formation of transcriptional condensates depends on RNA concentration. Moreover, the simulation results showed for the first time that different transcription factors may be selectively incorporated into the condensates depending on RNA concentration.
The RNA model developed here can be applied to a wide variety of intrinsically disordered proteins and is very useful for understanding molecular interactions in biomolecular condensates. The model is expected to contribute to the understanding of diverse life phenomena involving condensates and research on therapeutic means for neurodegenerative diseases such as Alzheimer's disease. The proposed computational method can be applied to biology and materials engineering and is expected to have spillover effects on interdisciplinary fields such as biophysics, physics, biology, and engineering.
Journal Information
Publication: Journal of Chemical Theory and Computation
Title: Coarse-Grained Model of Disordered RNA for Simulations of Biomolecular Condensates
DOI: 10.1021/acs.jctc.4c01646
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




