In vertebrate retinas, cone cells (the photoreceptors responsible for color vision) are arranged in a mosaic pattern. A research team led by Professor Ichiro Masai and former PhD Student Dongpeng Hu from the Developmental Neurobiology Unit at the Okinawa Institute of Science and Technology Graduate University (OIST) has discovered that a protein called Dscamb enables color-sensing cells in zebrafish retinas to maintain optimal spacing between cells through its action as a "self-avoidance enforcer," where cells avoid others of the same type, thereby achieving optimal vision. Their findings were published in Nature Communications.
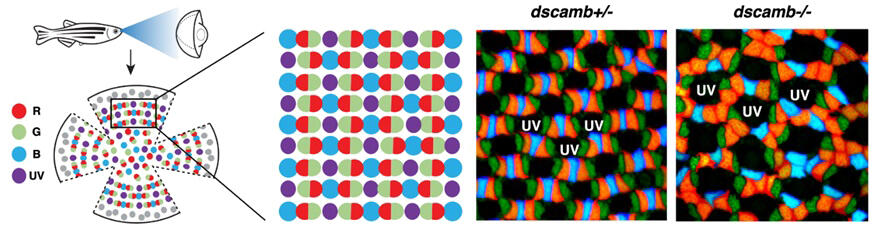
(Right panels) Cone mosaic pattern in fish lacking the dscamb gene on either or both of homologous chromosomes. Dscamb has been identified as the molecule that regulates red cone cell arrangement. Loss of Dscamb causes red cone clustering, leading to disruption of the regular cone mosaic pattern. Hu et al., 2025
Provided by OIST
In zebrafish, four types of cones combine to form a regular lattice-like cone mosaic pattern. Although such complex cone mosaic patterns have been reported in fish since the late 19th century, the molecules that directly control cone mosaic pattern formation had not been identified in any vertebrate until now.
Down Syndrome Cell Adhesion Molecule (DSCAM) is a protein that helps neural circuits form proper neuronal connections during development. DSCAM was first discovered on human chromosome 21, which is associated with Down syndrome. DSCAM proteins exist in many animals and help neural cells form circuits without becoming entangled. Zebrafish have three variants of this protein: Dscama, Dscamb, and DscamL1. Among these, only Dscamb is present in the photoreceptor cells of developing zebrafish eyes.
Hu said, "Since DSCAM regulates the self-avoidance mechanism in neural system development, we modified zebrafish genes to create specimens lacking functional Dscamb protein in this research. This allowed us to test our hypothesis that this protein is involved in cone mosaic formation. We discovered that in zebrafish dscamb mutants, the cone mosaic pattern, particularly the arrangement of red cones, was disrupted."
It has been reported that in the early stages of zebrafish photoreceptor differentiation, cone photoreceptor cells extend thin protrusions called filopodia from their apical regions, but the physiological role of these in photoreceptor differentiation was unknown. To clarify the role of Dscamb in cone mosaic formation, the research team visualized Dscamb protein in cells using fluorescent tagging techniques. They found that Dscamb protein was localized in the apical region, including the tips of the filopodia-like protrusions of cone photoreceptor cells.
The research team examined the behavior of filopodia in red cones. Time-lapse imaging revealed that in wild-type zebrafish, red cones extend filopodia toward adjacent red cones, make temporary contact, and then retract. However, no retraction of red cone filopodia was observed upon contact with non-red cones. This dynamic process gradually establishes appropriate spacing between red cone cells. In dscamb mutants, however, the filopodia of red cone cells failed to properly retract after contact with the same type of red cone cells, remaining attached to the surface of the apical region of adjacent red cone cells or even invading the region. As a result, red cone cells formed abnormal clusters, disrupting the regular mosaic pattern. This suggests that filopodia extending from the apex of cones function as antennae that explore the surrounding environment and sense whether nearby cones are of the same type. When filopodia extended from red cone cells have contact with other red cone cells, Dscamb proteins interact, triggering a repulsive reaction that causes the filopodia to retract. This self-avoidance mechanism allows red cone cells to maintain appropriate spacing from each other.
This self-avoidance mechanism is observed only between cone cells of the same type. Red cone cells recognize other red cone cells and show self-avoidance reactions. Similarly, blue cone cells recognize and avoid other blue cone cells. The research team discovered that Dscamb controls the spacing between red cones. On the other hand, the mechanism controlling spacing between blue cones is thought to exist separately from Dscamb. In other words, Dscamb functions as a sensor that recognizes red cones in the zebrafish cone mosaic formation process.
Masai said, "Computer analysis and modeling in this study confirmed that the mechanism of recognition and repulsion between cells of the same type can explain the observed cone mosaic patterns. This is the first identification of a molecular mechanism that directly controls cone mosaic formation in a species, opening up new possibilities for understanding similar processes in other vertebrates."
This discovery demonstrates the molecular basis for maintaining precise spacing of photoreceptors, which is essential for optimal vision, and provides a starting point for elucidating similar mechanisms in human retinal diseases.
Journal Information
Publication: Nature Communications
Title: Dscamb regulates cone mosaic formation in zebrafish via filopodium-mediated homotypic recognition
DOI: 10.1038/s41467-025-57506-1
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




