A research group led by Professor Chiba Chikafumi of the Institute of Life and Environmental Sciences at the University of Tsukuba has announced the successful development of an all-in-one Cre-loxP vector that enables the creation of Cre-loxP organisms in a single step (one introduction). These organisms allow precise control and modification of gene expression. The group discovered that placing a short DNA sequence named TAx9 immediately before the promoter region that expresses the Cre gene prevents unintended recombination in E. coli vectors. They also confirmed that this technology can be applied to newts and mice. The achievement is expected to be useful for a wide range of life science research. The results were published in the international academic journal Communications Biology on March 6.
Provided by the University of Tsukuba
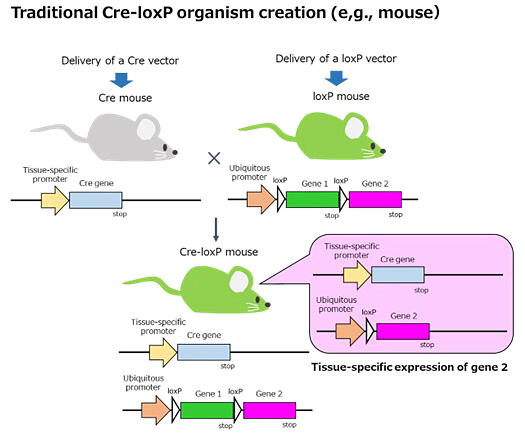
Organisms with the Cre-loxP system (Cre-loxP organisms) utilize site-specific recombination that occurs when Cre (a DNA recombination enzyme) acts on specific DNA sequences called loxP. This experimental system is widely used in life science research. Previously, Cre-loxP organisms were created by breeding Cre strains containing the Cre gene in the genome with loxP strains, as it was difficult to create all-in-one Cre-loxP vectors. When attempting to create an all-in-one vector, the Cre gene within the vector would spontaneously express in the E. coli used for vector synthesis. The problem with the conventional method is the occurrence of recombination with the loxP region within the same vector.
The research group had been attempting to create an all-in-one vector for tracking retinal pigment epithelial cells in adult Japanese fire-bellied newt retinal regeneration research, but had not succeeded. Meanwhile, they confirmed that they could create an all-in-one Cre-loxP vector for tracking skeletal muscle cells.
Provided by the University of Tsukuba
In this study, they analyzed in detail the tissue-specific promoter (African clawed frog-derived myofiber-specific promoter) used in the all-in-one Cre-loxP vector for tracking skeletal muscle cells.
As a result, they found that a DNA region of unknown function rich in AT/TA sequences upstream of the basic promoter involved in gene transcription activity was preventing spontaneous DNA recombination reactions in E. coli.
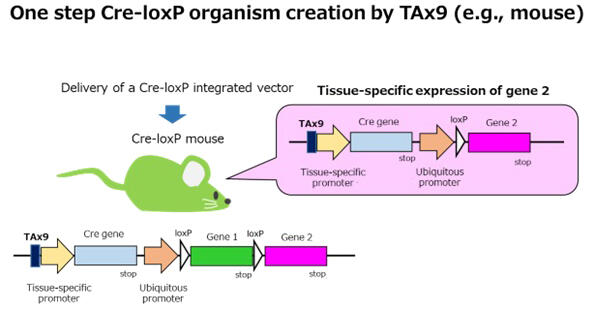
The team then identified the optimal AT/TA sequence for creating all-in-one Cre-loxP vectors and named it TAx9. By placing TAx9 immediately before the promoter region that expresses the Cre gene, they successfully synthesized an all-in-one Cre-loxP vector that prevents Cre expression in E. coli.
Furthermore, by applying this technology to mice and newts, they were able to create Cre-loxP newts that specifically fluorescently label retinal pigment epithelial cells and Cre-loxP mice that specifically fluorescently label skeletal muscle cells in a single step.
Chiba commented: "Cre-loxP organisms are very useful for investigating how genes function in living organisms. This technology (named TAx9 technology), which was accidentally discovered during regeneration research on Japanese fire-bellied newts, allows us to obtain Cre-loxP organisms at unprecedentedly low cost and in a short period of time, which is expected to greatly contribute to the advancement of life sciences. TAx9 technology can not only be combined with various other systems that manipulate gene function but is also believed to be applicable to a wide variety of species."
Journal Information
Publication: Communications Biology
Title: One-step Cre-loxP organism creation by TAx9
DOI: 10.1038/s42003-025-07759-9
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




