Associate Professor Keisuke Fukunaga of the Institute for Tenure Track Promotion at the University of Miyazaki, Assistant Professor Takamasa Teramoto and Professor Yoshimitsu Kakuta of the Faculty of Agriculture at Kyushu University, Professor Yohei Yokobayashi of the Nucleic Acid Chemistry and Engineering Unit at Okinawa Institute of Science and Technology (OIST), Professor Tomoaki Matsuura of the Earth-Life Science Institute (ELSI) at Institute of Science Tokyo, and their colleagues announced that they have elucidated the differences in binding mechanisms between two artificial RNA-protein complexes (RNPs) using X-ray crystallographic analysis. They successfully developed high-performance riboswitches (RNA switches for gene expression control) utilizing these RNPs and demonstrated their high performance in a cell-free protein synthesis system. Their findings were published in the international scientific journal Nucleic Acids Research on March 23.
Provided by the University of Miyazaki
Cell-free protein synthesis systems are being researched worldwide as tools for biochemistry research and for large-scale synthesis of specific proteins for medical applications. mRNA-binding proteins (RNPs) play a central role in the process where mRNA is transcribed from DNA in the nucleus and translated by ribosomes.
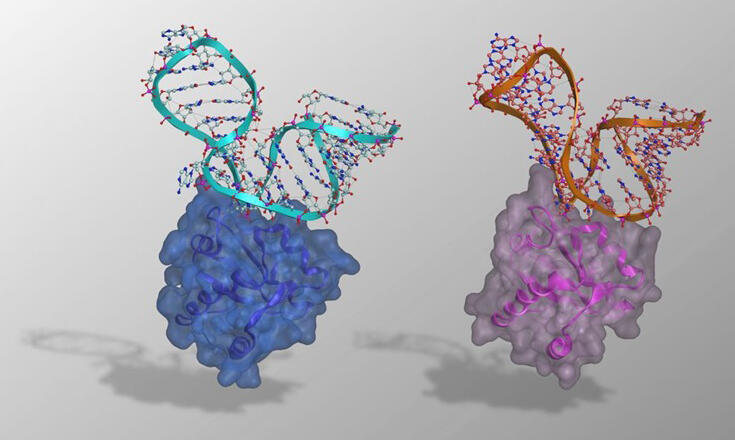
Previously, the OIST research group developed a PD-SELEX method to modify RNA-RBP binding mechanisms. By modifying the binding selectivity of archaeal L7Ae protein and its binding kink-turn RNA, which are commonly used as molecular parts in synthetic biology research, they successfully obtained two orthogonal RNA-RBP pairs: CS1-LS4 and CS2-LS12. However, the mechanism by which these highly homologous RNA-RBP pairs could form specific pairs remained unclear.
Therefore, in this study, they aimed to elucidate the interaction mechanism through X-ray co-crystal structure analysis. As a result, they successfully determined the structures of CS1-LS4 and CS2-LS12. They found that these two RNPs have overall structures very similar to their ancestral kink-turn RNA-L7Ae complex.
They also pursued the development of protein ligand-responsive riboswitches that function in a cell-free transcription-translation system.
They designed a system where RNA aptamer sequences (CS1 or CS2) were inserted immediately after the T7 promoter sequence, and when protein ligands (LS4 or LS12) are absent during transcription, the ribosome recognition sequence is blocked, inhibiting the translation of downstream genes. They created an ON switch where protein synthesis occurs only when protein ligands are present.
To verify the performance of these switches, the group conducted screening using the expression level of a fluorescent protein (EGFP) reporter as an indicator and confirmed that these formed a world-class high-performance cell-free riboswitch with an ON/OFF ratio equivalent to 500. They noted that an ON/OFF ratio in the three-digit range is unprecedented.
Furthermore, using the two developed riboswitches, they constructed a logic circuit (AND circuit) that controls the functional expression of γ-hemolysin and confirmed that it functions properly.
In the future, they plan to further improve the ON/OFF ratio and verify whether the RNA-RBP pairs function on different platforms such as E. coli.
Fukunaga commented: "We have successfully elucidated the structure of ultra-high affinity RNA-protein (RNP) pairs obtained through artificial evolution (PD-SELEX) and developed high-performance translation switches for cell-free systems using RNP parts. In the future, we would like to work on developing RNA devices that function in cells."
Journal Information
Publication: Nucleic Acids Research
Title: Structural insights into lab-coevolved RNA-RBP pairs and applications of synthetic riboswitches in cell-free system
DOI: 10.1093/nar/gkaf212
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




