A research group led by Professor Shinya Kuroda of the Graduate School of Science and Assistant Professor Keigo Morita from the Graduate School of Science's Molecular Genetics Research Laboratory at the University of Tokyo, in collaboration with Niigata University, Nara Institute of Science and Technology, the Medical Institute of Bioregulation at Kyushu University, and the Institute for Advanced Biosciences at Keio University, has revealed the pathological features caused by obesity using mice. They discovered that while starvation adaptation in the mouse liver is structurally robust against obesity, it is temporally vulnerable. These findings are expected to contribute to understanding the pathology of obesity and diabetes and developing treatment methods. The results were published in the international scientific journal Science Signaling on April 22.
Provided by the University of Tokyo
During starvation, numerous molecules must be mobilized in a temporally ordered manner to maintain metabolic homeostasis until the next meal. However, the design principles that enable this regulation and the pathological changes caused by obesity were not well understood.
In this study, the researchers focused on the liver, which plays a central role in metabolism. They comprehensively measured water-soluble metabolites (such as carbohydrates and amino acids), lipids, free fatty acids, acylcarnitines, acyl-CoAs, transcripts, proteins, and protein phosphorylation in the livers of normal mice and obese model mice (mice lacking leptin, an appetite-suppressing hormone) during starvation, and extracted molecules that showed significant changes.
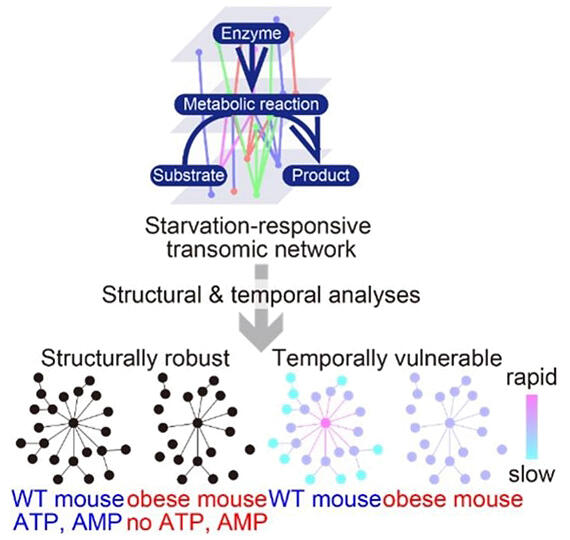
Using databases, they identified the metabolic reactions controlled by these extracted molecules and constructed a metabolic trans-omics network of mouse liver during starvation by connecting these reactions.
To organize the vast amount of difficult-to-analyze information, they interpreted this network by applying network theory and time constants (indicators of response speed).
The results showed that in healthy mouse livers, a scale-free-like network (a network consisting of a small number of hubs and many leaves) was constructed centered around energy-related metabolites such as ATP and AMP. In obese model mice, although ATP and AMP were replaced by lipids and other molecules, the scale-free-like network structure was maintained, demonstrating that the network structure is robust against obesity.
However, analysis using time constants revealed that this network is vulnerable against obesity. In normal livers, molecules at the center of the network, such as ATP and its metabolite AMP, responded more rapidly, and glycogen breakdown, gluconeogenesis (metabolic reactions that synthesize glucose from metabolites such as amino acids), and fatty acid breakdown responded in an orderly manner. In contrast, this order was lost in obese model mice, indicating vulnerability against obesity.
Kuroda commented: "Recently, it has become possible to obtain vast amounts of data in biology as well. However, it is not easy to understand this as a system rather than just using it for screening. In this study, I believe we succeeded in capturing the mechanism of starvation adaptation, which is extremely important for biological survival, as a system by merging basic network theory with biology."
Journal Information
Publication: Science Signaling
Title: Structural robustness and temporal vulnerability of the starvation-responsive metabolic network in healthy and obese mouse liver
DOI: 10.1126/scisignal.ads2547
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




