Associate Professor Fumiaki Ohtake, Graduate Student Mai Morita, Undergraduate Student Miyu Takao, and Graduate Student Ryotaro Chiba of Hoshi University, in collaboration with Associate Professor Yusuke Sato and Graduate Student Honoka Tokuhisa from Tottori University, Professor Yasushi Saeki from the University of Tokyo, and their colleagues, have announced the discovery of a new principle of antagonism relating to protein degradation within cells. They have solved the seemingly paradoxical mystery of how protein degradation is achieved amid antagonism between enzymes that add and cleave ubiquitin in cells. They also found that this enzymatic antagonism regulates inflammatory responses. This finding is expected to lead to the development of new treatments for inflammatory diseases. Their results were published in the international scientific journal Nature Communications on March 24.
Provided by Hoshi University
Proteins synthesized in cells are degraded when they become unnecessary, but if degradation does not occur properly, unnecessary proteins accumulate in cells, causing diseases such as cancer and neurodegenerative disorders. Conversely, if proteins are excessively degraded, cells lose their function, which can also cause disease.
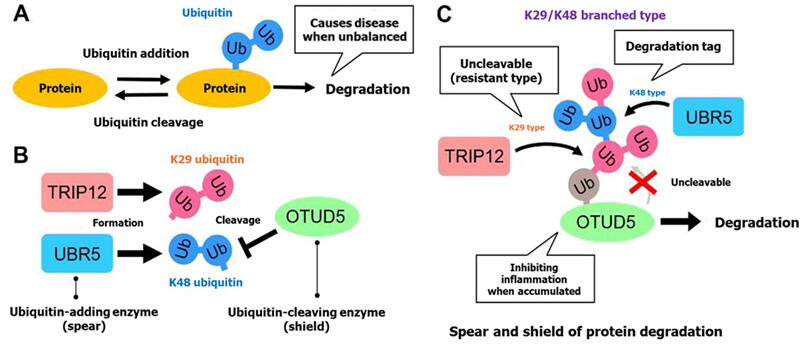
The main pathway responsible for protein degradation in cells is the ubiquitin-proteasome system, where ubiquitin, a marker (protein), is attached to unnecessary proteins, and these ubiquitin-tagged proteins are transported to an enzyme complex called the proteasome to be degraded. Within cells, there are numerous enzymes that add and cleave ubiquitin, creating an antagonistic relationship. However, it remained unclear how protein degradation occurred amid this cellular antagonism.
The research group focused on the ubiquitin-adding enzymes TRIP12 and UBR5. Added ubiquitin forms ubiquitin chains where ubiquitin molecules are linked together in a chain-like structure, serving as a marker for degradation. TRIP12 forms "K29-type" chains and UBR5 forms "K48-type" chains, which are different types of ubiquitin chains. The researchers discovered that the ubiquitin-cleaving enzyme OTUD5 is a common degradation target for both.
Through investigation of the mechanism, they found that OTUD5 easily cleaves K48-type chains, preventing UBR5 alone from promoting degradation. In contrast, K29-type chains cannot function as degradation tags by themselves but are resistant to OTUD5 and avoid being cleaved. When TRIP12 and UBR5 coexist, they form "K29/K48 branched ubiquitin chains" that both evade degradation by OTUD5 and function as tags. They also discovered that OTUD5 is an inhibitory factor of inflammatory responses triggered by inflammatory cytokines, and when OTUD5 accumulates due to lack of degradation control by TRIP12/UBR5, cellular inflammatory responses are suppressed.
Ohtake commented: "This research pursues the fundamental mechanism behind the active degradation of unnecessary proteins in cells. This knowledge could potentially be applied to 'targeted protein degradation drug discovery' technology that artificially degrades disease-causing proteins in the future."
Journal Information
Publication: Nature Communications
Title: Combinatorial ubiquitin code degrades deubiquitylation-protected substrates
DOI: 10.1038/s41467-025-57873-9
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




