Associate Professor Kan Shoji from the Department of Mechanical Engineering, Faculty of Engineering, and Graduate Student Hiromu Akai from the Graduate School of Engineering, Advanced Engineering Program, Nagaoka University of Technology announced the joint development of a DNA nanopore sensor capable of selective molecular detection with Associate Professor Takuya Mabuchi from the Institute of Fluid Science, and Graduate Student Taichi Hirano from the Graduate School of Engineering, Tohoku University. The sensor has potential applications for various biomolecules through modifications of the base sequences of the aptamers that constitute the sensor. This development is expected to contribute to drug discovery and disease diagnosis. Their results were published in the electronic edition of the international academic journal Small on May 2.
Provided by Nagaoka University of Technology
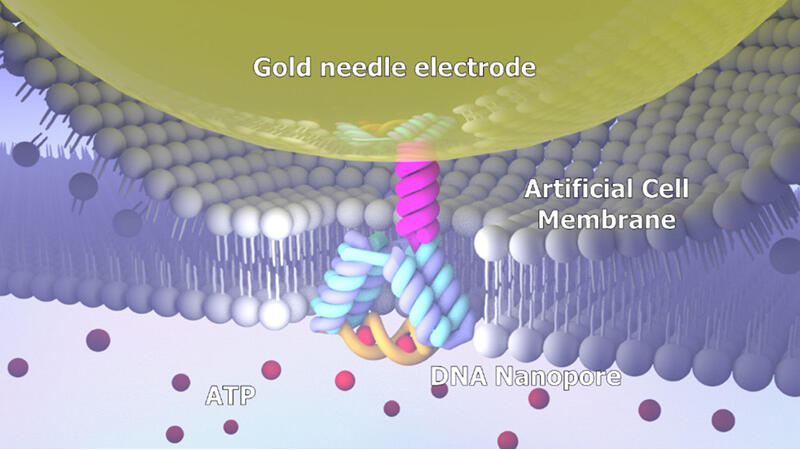
Nanopore sensors are molecular measurement technologies that detect target molecules through changes in ionic current caused by interactions between nanometer-scale pores constructed on thin films and target molecules. They are used for DNA sequence analysis and protein detection. In principle, various molecules passing through nanopores can be detected, so custom-made nanopores are required to detect only target molecules. In recent years, the application of "DNA nanopores," which are constructed from DNA and allow flexible control of size, shape, and functionality, to these sensors has been anticipated. Although DNA nanopores capable of inducing opening and closing actions in response to molecules have been reported, characteristic evaluations regarding molecular selectivity and temperature dependence, which are essential for sensor applications, had not been conducted due to low insertion efficiency into artificial cell membranes.
Therefore, the research group developed a DNA nanopore sensor that dynamically opens and closes in selective response to ATP (adenosine triphosphate) using structural DNA nanotechnology.
The DNA nanopore developed consists of a transmembrane domain that functions as an ion pathway in artificial cell membranes and ATP-binding aptamers (nucleic acid molecules that bind to specific molecules and form three-dimensional structures), which are nucleic acid molecules with the function of selectively binding to the target molecule adenosine triphosphate (ATP).
Next, using their unique "DNA nanopore probe technology," the researchers inserted the DNA nanopores into artificial cell membranes. By measuring ionic current, they successfully observed the opening and closing actions of DNA nanopores in response to ATP. They also found that the proportion of closed states (where aptamers and ATP are bound) changes depending on ATP concentration.
Furthermore, by using molecular dynamics simulations that can observe structural changes at the atomic level, they successfully clarified the detailed ATP-responsive opening and closing mechanisms of DNA nanopores.
Shoji commented: "In this research, we reported the world's first nanopore sensor using molecule-responsive DNA structures. It possesses high sensitivity, high selectivity, and high temporal resolution. Furthermore, by designing aptamer sequences, we can tailor-make molecular selectivity, so we expect it to become a biosensor applicable to drug discovery and cell observation."
Journal Information
Publication: Small
Title: Specific ATP Detection Using Molecule-Responsive DNA Nanopores
DOI: 10.1002/smll.202409293
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




