On May 20, Shimadzu Corporation launched "MicrobialTrack," microbial identification software that utilizes a large proprietary database, as a cloud service both domestically and internationally. This software is designed for microbial identification in pharmaceutical testing, food hygiene management, and river water quality inspection. The suggested retail price starts from 1.32 million yen (tax included). The sales target is 100 licenses in total domestically and internationally within one year of release.
Provided by Shimadzu Corporation
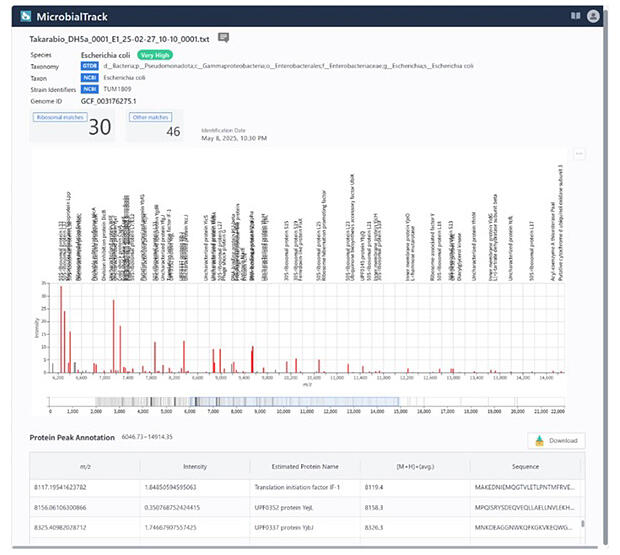
The new product references databases with microbial measurement results obtained by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS). It identifies which of approximately 85,000 bacterial and archaeal species —including published classifications based on the International Code of Nomenclature of Prokaryotes, as well as difficult-to-culture and uncultured classifications—the target organism corresponds to within 3 hours.
Microbial identification is experiencing growing demand worldwide due to increasing hygiene awareness, the spread of chilled foods, and frequent occurrences of drug-resistant bacteria. For clinical and food microbial testing, there is a "fingerprint method" that identifies organisms based on pattern similarity with measurement results obtained from MALDI-TOFMS.
Although testing only takes about 3 hours, application has been limited to specific purposes due to the database size being restricted to 5,000 known classifications and accuracy issues with identification.
As a result, "genetic analysis methods" have been used for investigating unknown microorganisms. This involves extracting DNA from microorganisms and decoding the base sequences of genes. However, identification takes approximately 2 days and requires specialized knowledge, so it is often outsourced to contract analysis organizations. In contrast, the new product "MicrobialTrack" adopts a "proteomics approach " that identifies organisms based on similarity to theoretical masses of microbial proteins estimated from genome sequences.
The theoretical mass database for prokaryotic microbial proteins "GPMsDB" used by the product contains theoretical masses of prokaryotic microorganisms derived from approximately 400,000 data entries obtained from public genome databases.
In addition to published microorganisms, 85,000 species including uncultured and difficult-to-culture organisms without assigned scientific names are included as analysis targets. This enables analysis of a wide range of microorganisms that were not previously covered by MALDI-TOFMS. Furthermore, hundreds of thousands of new microbial genome data entries are registered annually in public genome databases. "GPMsDB" is updated accordingly, allowing microbial queries to be implemented using databases that are always based on the latest information.
"MicrobialTrack" is a cloud service and does not require installation of dedicated software or equipment. Analysis work can begin simply by entering an ID and password and logging into the website.
Analysis results are displayed in ranking format showing microorganisms with high similarity, along with color-coded icons corresponding to reliability levels. Since MALDI-TOFMS analysis results and protein information based on proteomics (comprehensive protein analysis) are also provided, even specialized analyses requiring expert knowledge can be performed easily.
During the license period, microbial identification using the latest "GPMsDB" will be possible without additional costs. It will also support measurement results obtained from MALDI-TOFMS instruments from manufacturers other than Shimadzu.
This new product was developed based on joint research results with the National Institute of Advanced Industrial Science and Technology (AIST) and the National Institute of Technology and Evaluation (NITE). AIST and Shimadzu in 2023 announced the theoretical mass database for prokaryotic microbial proteins "GPMsDB" and microbial identification methods using proprietary algorithms for analyzing mass spectrometry results.
MALDI is the technology that earned Koichi Tanaka, Executive Research Fellow at the company, the Nobel Prize in Chemistry. It is a technique that ionizes high-molecular compounds such as proteins without breaking them by irradiating laser light on samples mixed with a matrix (ionization auxiliary agent).
This article has been translated by JST with permission from The Science News Ltd. (https://sci-news.co.jp/). Unauthorized reproduction of the article and photographs is prohibited.




