Yasuo Suda, President, SUDx-Biotech Corp. and Professor, Graduate School of Science and Engineering, Kagoshima University
Overview
SUDx-Biotech Corp. was founded in September 2006 as an authorized venture of Kagoshima University to provide our technology to the world, based on the achievements of a pre-venture project, led by myself under the Japan Science and Technology Agency (JST), focusing on developing practical applications for sugar chips between October 2003 and September 2006. Apparently, it was the 51st authorization under the JST.
The surfaces of our cells are covered with a diverse range of sugars sized in the nanometer range called glycans, or sugar chains. Sugar chains interact with specific proteins or other sugar chains to communicate information to cells as a part of biological reactions, such as immunity responses. Glyco-sciences such as glycobiology are also gaining focus within the life sciences after it was discovered that they are related to carcinogenesis and viral infection in cells. However, a great deal of funding and effort is required to identify sugar chains with a clear structure, a requirement for research at the molecular level, often making it impossible to proceed with research. To solve this problem and dramatically advance glycoscience research, we have developed two tools, the sugar chip and the sugar chain-immobilized gold-nanoparticle, that immobilize sugar chains with well-defined structures on gold chips sized at the nanometer scale. Furthermore, we found that these tools can be applied to capture, concentrate, and filter viral particles, and we succeeded in developing rapid and highly precise testing methods, combined with polymerase chain reaction (PCR) testing, for viruses that infect humans and livestock, such as influenza, norovirus, herpes, and porcine epidemic diarrhea. We have also developed an in vitro diagnostic kit based on this technology for the novel coronavirus (SARS-CoV-2) that caused the pandemic in 2020. The SUDx SARS-CoV-2 detection kit received insurance coverage in June 2020, and we subsequently began the sale in December 2020 of our SGNP nCoV/Flu PCR testing kit, a non-invasive test that detects both SARS-CoV-2 and influenza from saliva specimens, after it received pharmaceutical approval.
About influenza
We are taught basic anti-viral preventive measures from early childhood to help prevent the spread of influenza, such as wearing masks, hand washing, avoiding crowded areas, vaccination, and building up the immune system, and these are generally the same measures for preventing COVID-19. Despite this, we wondered why it was that influenza spread through the population every year, and that question is what motivated us to apply our technology to a diagnostic testing kit.
Laboratory diagnosis of influenza in Japan commonly consists of immunochromatographic testing of antigenic proteins. Unfortunately, that approach is not adequately sensitive, and a positive result can often only be obtained more than 24 hours after the onset of symptoms. Meanwhile, the test specimens typically consist of invasive nasal swabs which contain a large number of viral antigens. There is no shortage of patients who went through the painful diagnostic process, only to test negative, but to then go on to develop more severe symptoms. The trauma of that experience leads people to avoid getting tested in the future, and the disease can worsen accordingly when left untreated with no influenza antiviral drugs administered.
To solve this problem, we developed a highly sensitive and painless test. A video has been made available on our website (A Painless PCR Test for Influenza Using Saliva | SUDx-Biotech Corp.) offering the best explanation of the method, but I will also provide a brief explanation here as well.
The surfaces of cells are covered with sugar chains, and this prevents viruses from seeing the proteins within the cell cortex. Therefore, the viruses first attach themselves to the sugar chains. Then, the viruses invade the cell and infect it as true receptors (which is believed to be a sialic acid-containing sugar chain in the case of influenza). As shown in Fig. 1, we first identified the sugar chains that the viruses adhere to using a sugar chip. We then immobilized the sugar chains in gold nanoparticles to form SGNPs (sugar chain-immobilized gold-nanoparticles) that are smaller than the virus. When SGNPs are added to a solution containing the virus, they adhere to and trap them. Since the captured virus is heavier, a precipitated fraction containing virus particles can be obtained using even a low-speed centrifuge. By either adding a surfactant to the solution or by adding water and heating it, the viral particles are destroyed and release RNA (ribonucleic acid). The solution has already been partially purified when collecting the viral particles, so the viral RNA that is released is of adequate purity to be detected via RT-PCR (reverse transcription-polymerase chain reaction). Magnetic separation can be achieved by making the nanoparticles magnetic, so we devised a method to add magnetic microparticles to guide the process, which enabled us to obtain an RNA solution for RT-PCR testing in just around three minutes after obtaining samples.
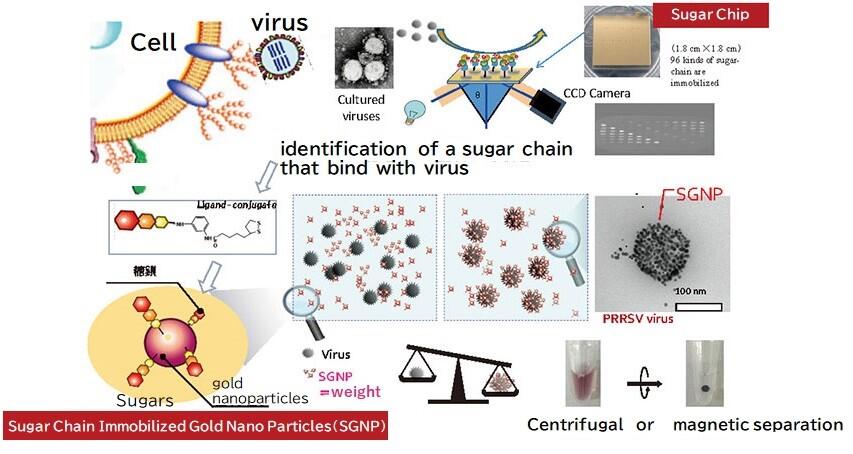
Table 1 shows a portion of the first clinical results we received with our testing method. The data is a comparison of the test results from nasal swabs and a rapid diagnostic kit (immunochromatography method) used on patients with symptoms of influenza at small clinics and university hospitals in Kagoshima, and the RT-PCR test results carried out after extracting RNA from roughly 0.5 mL of saliva from the same patients using the SGNP method (hereinafter, SGNP/PCR method). The comparison showed that the SGNP/PCR method had an overwhelmingly superior positivity rate (sensitivity). We found, to our surprise, that over 50% of patients with influenza symptoms who tested negative using the rapid diagnostic kit went on to test positive with the SGNP/PCR method. Subsequently, after five years of continuous clinical research, our method gained approval from the Advanced Medical Care Council of the Ministry of Health, Labour and Welfare in the Advanced Medical Care A category in 2017, under the title "Support for infectious disease treatment and nosocomial infection control via a highly sensitive virus testing method using sugar chain nanotechnology. "
The main objective of this Advanced Care A study was to determine the time period after the onset of disease when the SGNP/PCR method performed best compared to rapid diagnostic kits, something previously unknown. A survey carried out in Hospital I during the 2018-2019 flu season found influenza in the saliva of 17 of a total of 46 patients with influenza symptoms who were given the rapid diagnostic test within 24 hours of developing symptoms and tested negative, including influenza type A (9 people), B (7 people), and both A & B (1 person). Meanwhile, it was found in the saliva of 64 out of 194 patients with influenza symptoms who tested negative at Hospital M (53 with type A, 9 with type B, and 2 with both). This demonstrated that our method was an effective test even at less than 24 hours from the onset of symptoms. Furthermore, dilutions of saliva from 30 patients were subjected to virus isolation culture experiments using MDCK cells, and both the accuracy (positive match rate, 13/13) and specificity (negative match rate, 17/17) were in perfect agreement, indicating that the SGNP/PCR method is capable of measuring infectious viruses.
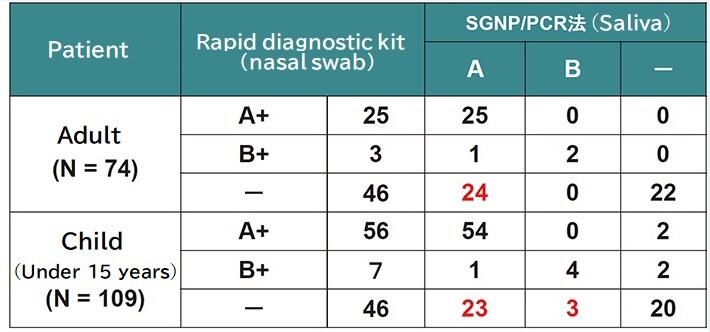
A comparison between the results of the rapid diagnostic test and the SGNP/PCR test using saliva.
Based on these results, we then conducted a clinical performance test in consultation with the Pharmaceuticals and Medical Devices Agency (PMDA). As there has been no case in which saliva was used as a sample for any hitherto approved in vitro diagnostic test for influenza, virus isolation culture using MDCK cells from the nasal secretions of the same patients were used as a control for the test of the SGNP/PCR method using patient saliva. The quantity of viral material in nasal secretions has been reported as being greater in numerous papers, so there was some concern as to whether we could obtain a 90% or greater testing rate in comparison. Nonetheless, we began our clinical testing at four clinics and hospitals in Kagoshima in December 2019. However, we were able to collect almost no specimens from March 2020 due to the effect of the COVID-19 pandemic, so we instead compared our data with that of 463 patients for whom all data, including medical questionnaires, was available, with the permission of the PMDA. The comparison luckily showed an overall 95% match rate, with an accuracy of 93% and specificity of 95%, resulting in the successful completion of the clinical study.
COVID-19
In February 2020, a patient suffering from a novel coronavirus infection was admitted to the Hamamatsu Medical Center at which Dr. Tajima, a joint researcher under Advanced Medicine A, works. While we had no experience testing for human coronaviruses to date, we knew that we could concentrate porcine coronaviruses (the porcine epidemic diarrhea virus) using SGNP, the same approach as we use for the influenza virus, and we began clinical research on SARS-CoV-2 in consultation with Dr. Tajima. Dr. Tajima collected two types of specimens: saliva and throat or nasal swabs. RNA was then extracted from the swabs using the Qiagen method at the Hamamatsu Environmental Health Center using the protocol established by the National Institute of Infectious Diseases (NIID), and PCR testing was performed. Meanwhile, the saliva samples were concentrated using a kit for influenza, and RNA was extracted and sent to us in Kagoshima. We then carried out a PCR test using a partially modified primer probe recommended by the US CDC (Centers for Disease Control and Prevention) and the NIID. We found that SARS-CoV-2 remained in the patient's saliva up until 35 days after hospitalization, and the patient was released from the hospital on the 42nd day once the saliva test and nasopharyngeal swab test were both negative. This study suggested that it would be possible to obtain the same test results as the nasopharyngeal swabs by testing the amount of saliva excreted as soon as one wakes up in the morning.
Following this, we continued our clinical research using both saliva and nasopharyngeal swabs from patients suspected of having COVID-19 at the Hamamatsu Medical Center. Accordingly, we evaluated our SGNP/PCR method using the test results from nasopharyngeal swabs at the Hamamatsu Medical Center as a control. The results showed a 100% match with accuracy (10/10) and specificity (15/15) for the nasopharyngeal swabs, and a 90% match for accuracy (9/10) and 100% match for specificity (15/15) for saliva. These results were explained to the Ministry of Health, Labour and Welfare, and the agent was approved for insurance coverage as of June 10, 2020, as the "SUDx SARS-CoV-2 detection kit" for emergency use research reagent.
Application for pharmaceutical approval
After the clinical performance test for influenza exceeded 90% in terms of accuracy and specificity, we had a general consultation with the PMDA on June 24, 2020, to discuss applying for pharmaceutical approval (approval under the Act on Securing Quality, Efficacy and Safety of Products Including Pharmaceuticals and Medical Devices). We then received the opinion that there was no issue with case 463 and that we could apply. During the discussions, I mentioned that I had been working in my spare time on a testing methodology for measuring for both influenza and COVID-19 in saliva at the same time using a combination with the COVID-19 testing kit that had just received insurance approval. Then, It was recommended to me that I should apply for pharmaceutical approval for the simultaneous testing method instead, in preparation for the 2020 flu season. Accordingly, I developed a protocol using an artificial specimen that mixed inactivated SARS-CoV-2 with positive controls of both influenza A and B in saliva, a total of three viruses, which I explained at the general consultation two weeks later, and received the approval of the PMDA. A representative of the Ministry of Health, Labour and Welfare was also present at the consultation and requested that we consider mass production.
I then selected enzyme systems, probes, primers, and multiplex dyes for the simultaneous measurement of the three viruses, and subsequently tested a total of 120 artificial specimens. Each was tested separately for SARS-CoV-2, influenza A, and influenza B for control purposes. The results were 100% for both accuracy (105/105) and specificity (15/15). These results were compiled as the SGNP nCoV/Flu PCR Testing Kit and the application for pharmaceutical approval was submitted in September. I followed this by also testing artificial samples of nasal swabs and nasopharyngeal swabs in addition to the saliva test, finally receiving approval on October 23 for an in vitro diagnostic test for both COVID-19 and influenza using either saliva, nasal swabs, or nasopharyngeal swabs, and the price came under insurance coverage on November 10.
Ours is the only kit that can simultaneously test for influenza and COVID-19 using saliva, and we believe that no other kit has been officially approved at this time. Saliva is a sample which does not cause the patient any pain or discomfort, and samples can even be taken by the patient themselves for submission online.
A testing method that avoids false positives
The SGNP/PCR method collects viral particles at the start, so we considered that the quantity of virus measured could also describe the patient's state of illness. To prove this, we conducted a follow-up experiment on inpatients with COVID-19 infections. Referring to the example of Hamamatsu Medical Center described above, we collected saliva from patients admitted to Hospital T in Kagoshima City in the morning when the patients had just woken up. That saliva was mixed with the dilution solution used for the SUDx SARS-CoV-2 testing kit and stored at 4°C. RNA from SARS-CoV-2 was then extracted and filtered from the solution using the Qiagen method (Boom method), while viral material was also gathered using the SGNP method followed by RNA extraction. Both sets of RNA were then subject to both RT-PCR and PCR testing using the same protocol to determine the quantity of virus in the original solution.
Here, I will describe two different cases as an example. Patient TCH-59, in Fig. 2, was tested using both the SGNP method (blue bars on the graph) and the Qiagen method (grey bars on the graph) on the second day of hospitalization, and a very high concentration of RNA was found in both tests. Under the SGNP method, apart from a slight increase on the 5th day, the concentration continued to decline by the day, finally being undetected from the 6th day. In comparison, the RNA concentration detected by the Qiagen method remained largely unchanged until the 10th day. The patient's condition was improving by the day, and they became completely asymptomatic from the 7th day. Meanwhile, very little RNA was detected via the SGNP method in Patient TCH-61 on the second day of admission, while none was detected by the Qiagen method. This is likely because the specimen is concentrated by 25 times compared to only 2.5 times for the Qiagen method as a basic calculation. The SGNP method showed no change on day three, but a rise on day 4. In comparison, the Qiagen method detected the RNA on day three, and showed almost the same level on day 4. We were unable to collect specimen on days 5 and 6 due to it being a weekend, but the results on day 7 showed a rise in both the SGNP and Qiagen methods. The patient's condition worsened further, and they were transferred to a specialized hospital.
As demonstrated by patient TCH-59, the Qiagen method cannot differentiate between RNA from viral fragments and RNA from within viral particles, PCR testing carried out while they are recovering can result in a false positive, which may cause added confusion. For that reason, PCR testing is not commonly carried out for hospitalized patients. Even without testing, patients like TCH-59 can be discharged after 10 days of hospitalization if they do not exhibit any further symptoms. In comparison, both the Qiagen and SGNP methods produced data that predicted the worsening of symptoms in patient TCH-61. This suggests that the PCR testing that is consistent with the symptoms of the patient, such as the SGNP method, is extremely effective. There are many more such examples, which are currently being compiled into a paper. In both cases, the SGNP method results in an RNA quantity that can explain recovery from symptoms more so than the Qiagen method and enables a testing regimen with the potential for fewer false positives.
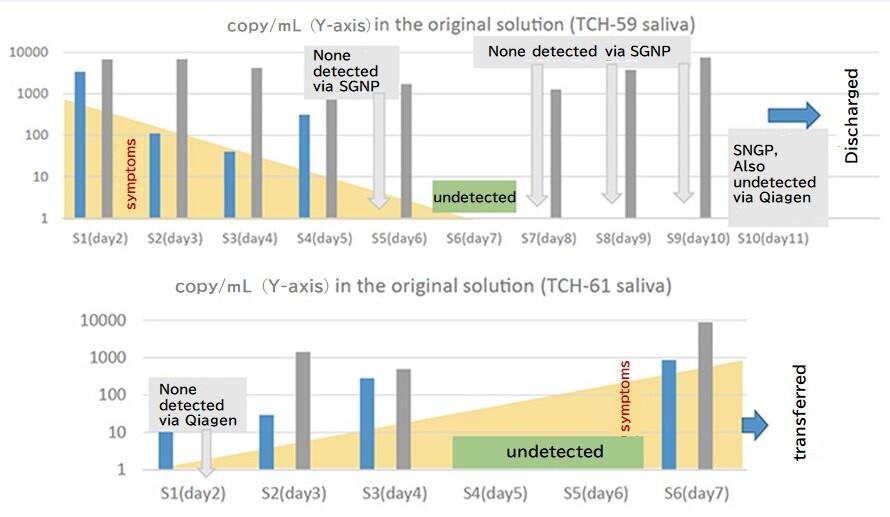
(blue bars: SGNP, gray bars: Qiagen.)
In closing
The SGNP method has the advantage of being simple and quick as a pretreatment method for PCR testing and enables PCR testing to accurately represent the state of illness by concentrating the viral particles and thereby increasing sensitivity. Some cases show that this method may enable patients in the convalescent stage in particular to return to their work early. Accordingly, the method is likely to be useful during the Tokyo Olympics and Paralympics for athletes who are infected.
We are currently working on the development of a multiplex compatible high-speed PCR testing device for use when rapid results are required for small numbers of specimen. We also intend to develop a device for automatically processing specimen using the SGNP method.




