Since ancient times, humanity has been fascinated by the organisms around us that are able to produce light. The mechanism behind luminescence has been clarified over the past few decades and this discovery is now being utilized across many research themes in the life sciences field. The evolution and development of these luminescence abilities had, however, remained unknown until now. Yuichi Oba, Professor of Environmental Biology, of the College of Bioscience and Biotechnology, Chubu University is one researcher who aims to shine a light on the unknown evolutionary pathways that allowed for the development of luminescence.
Through focusing on the enzymes related to the luminescence of fireflies, the posterchild for luminescent organisms, and the utilization of genetic engineering and computational science, Prof. Oba succeeded in the recreation of the light that was emitted by the fireflies of 100 million years ago.

Communication through flickering and utilization as a research tool
The mesmerizing light of fireflies has been a familiar sight in the early summer since long ago. (Fig. 1). They are commonly distributed in areas with high rainfall across tropical and temperate regions. There are around 2000 species that can be found throughout the world and around 50 of those inhabit the islands of Japan. The color of light emitted, and the related flashing patterns, vary between species. This begins from the larval stage of the life cycle, and continues in the adult phase in about half of firefly species.
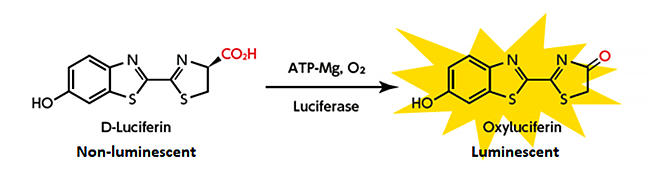
The emission of light is generally considered to be a form of communication between individual fireflies. The purpose of this varies between life phases, with it being involved in courtship behavior in adults and larval fireflies using it as an aposematic signal (warning) to predators of their poisonous characteristics. In the abdomen of adult fireflies, the enzyme luciferase catalyzes the oxidation of luciferin which results in the characteristic glow. This takes place in an organ known as the photophore which is present in most bioluminescent organisms (Fig. 2). Regardless of the color emitted, previous research has shown that luciferin is a common substance found in all species of firefly across the world. Luminescence is also extremely energy efficient, and the process essentially releases no heat, which has resulted in it being referred to as 'cold light'.
The understanding of luminescence resulting from recent research has led to the development of diagnostic tools utilizing its mechanism. These tools are widely used in research in the life sciences. If the intensity and color of the light emitted by the luminescent substances used in these can be manipulated freely it will lead to the development of more useful and user-friendly tools. The luciferase found in extant fireflies is now common knowledge, but the timing and acquisition of its luminescent capabilities is yet to be clarified.

Prof. Oba has had a fondness for insects since his childhood. Ever since he began studying luminescent organisms as a college student, he has been investigating earthworms, mushrooms, fishes, and other light-emitting organisms. Bioluminescence is not found in amphibians, reptiles, birds, and mammals, but it is a commonly seen phenomenon in fish and fungi. Regarding his research motives, Prof. Oba had this to say: "The acquisition of luminescence function is thought to have occurred independently in the evolution of each group and has occurred many different times in living organisms. Clarifying the process of its evolution in the firefly is a topic of great academic interest."
The evolution of similar enzymes that have different functions
Prof. Oba sought the origin of luciferase and so began his search by looking for genes with base sequences that resemble that of luciferase in insects that do not emit light. One organism that fits this bill is the model organism Drosophila, which had its whole genome sequenced for the first time in the early 2000s and when the genome database was investigated, a gene with a base sequence that closely resembles that of luciferase was found.
At the time of discovery, the function that this gene encodes was unknown, but it was later found to encode fatty acyl-CoA synthetase (ACS). This enzyme is involved in fatty acid metabolism and has a completely different role than the luminescent function seen in other insects.
Prof. Oba reminisces on those times "This enzyme as it is clearly does not cause luminescence, however, it has a base sequence very close to that which encodes luciferase. I believed that there was a high chance that the evolution of ACS was the origin of luciferase seen today. With that in mind I modified the amino acids of ACS to try to obtain the luminesce abilities seen in luciferase. As ACS is a protein composed of approximately 550 amino acids, if one were to change each encoded amino acid one at a time to different one, it would require a huge number of experiments to determine the mutations that resulted in luciferase.
Prof. Oba hypothesized that if changing a section of the amino acids led to luciferin glowing, then those amino acids were the likely luciferin binding site of luciferase. Using the three-dimensional structure of the bound luciferin-luciferase complex, he narrowed down the amino acid candidates for ACS conversion to seven. Furthermore, because four of the seven amino acids were the same as those of luciferase, the remaining three were left as the final candidates and as expected, when Prof. Oba changed the amino acids of ACS one by one, light was emitted. Although the light was extremely weak compared with that emitted currently by fireflies, Prof. Oba had proven that the origin of luciferase was ACS.
A self-deciphered ancestral sequence but specialized knowledge needed for confidence
Turning to the experts for help: "Wood from a woodcutter"
The genes of living organisms are written using four bases: adenine, guanine, thymine, and cytosine. The genes of organisms encoded by these bases mutate at random and change over time. Based on these changes it is possible to identify when two species diverged and differentiated by examining the differences present in their base sequences. Using gene sequences from multiple extant fireflies, Prof. Oba attempted to elucidate the genetic evolutionary process leading from ACS to the emergence of luciferase. Reminiscing on those days, Prof. Oba said, "I learned information biology on my own and tried to estimate the phylogeny, but I wasn't able to confirm if the results I got were correct. I realized that I needed to collaborate with phylogenetic experts. As they say, 'for wood, ask a woodcutter'." Although they had yet to meet, hoping that his published papers would be enough enticement and resolving to get his cooperation, Prof. Oba took a chance and sent an email to Prof. Tsuyoshi Shirai of the Department of Frontier Bioscience, Nagahama Institute of Biosciences. Prof. Shirai, who studied information biology at Nagoya University, was working on the structural analysis of proteins as well as on the phylogeny estimation of a protein called galectin that exists in the mucous membrane of conger eels.
"The estimation of ancestral gene sequences is scientifically important research, but I felt that it was difficult to convey the significance to the general public. Prof. Oba's research is clear and easy to understand; after all, anyone can understand why someone would ask how fireflies came to be able to glow. I felt that it had the power to appeal to the public at large," said Prof. Shirai, reflecting on his first impression of Prof. Oba's work. Prof. Shirai immediately decided to accept Prof. Oba's request, and that is how their joint research began approximately a decade ago.
Accumulating "plausibility"
Inferring ancestors from a molecular phylogenetic tree
One of the ancestral sequence estimation methods that Prof. Shirai proposed to Prof. Oba was the "maximum-likelihood method." Through the construction of molecular phylogenetic trees, which indicate when species diverged from a gene sequence, researchers can estimate the genes possessed by past organisms. The maximum-likelihood method estimates the 'most probable outcome' of a statistical analysis. In short, the observed outcome is the most likely result of the observed series of events. For example, when comparing results with a previous stage, the likelihood is high if the outcome is as close as possible to what was expected, whereas it is low if the outcome is an unexpected one. In the maximum-likelihood method of ancestral sequence estimation, sequences that have a high likelihood of having occurred are accumulated, and the ancestral sequence of many generations ago is estimated. "To date, many species have emerged and then become extinct, and so it is impossible to estimate past sequences from information on existing species alone with 100% accuracy. However, the more detailed the molecular phylogenetic tree is, the more accurate the estimation will be," said Prof. Shirai, explaining the difficulty of ancestral sequence estimation.
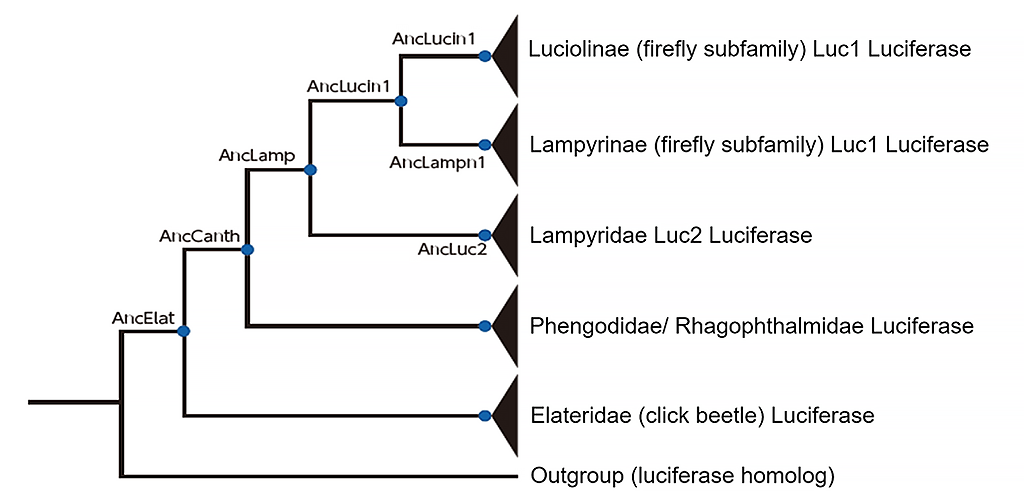
With the cooperation of Prof. Dai-ichiro Kato, who had also been studying the luminescence of fireflies, the group began the process of laying out the most detailed molecular phylogenetic tree as possible. On top of approximately 30 species of firefly, the group also collected and analyzed other families that are closely related, such as Rhagophthalmidae, Elateridae, and Omethidae and added their data to the phylogenetic tree. Fortunately, almost all subfamilies of firefly are present in Japan and so the species variation of the phylogenetic tree was sufficient despite the collection region being limited to this country. Additionally, in a joint study with the Escherichia coli Institute for Basic Biology and the Massachusetts Institute of Technology in the USA, Prof. Oba deciphered the entire genomes of the Japanese Heike firefly (Aquatica lateralis) and the American common eastern firefly (Photinus pyralis). The habitats of these two species are quite different but despite diverging approximately 105 million years ago, they share common genes for two types of luciferase: Luc1 and Luc2. Luc1 is used in larval and adult photophores, whereas Luc2 is expressed in the eggs and through the whole body of pupae.
Through this knowledge the team discerned that the two genes served different purposes. Given that these two relatively distantly related species have two luciferase genes, then there is a very high possibility that all firefly species share this characteristic. Prof. Oba and colleagues used the Luc1 and Luc2 genetic information to create a detailed molecular phylogenetic tree (Fig. 3). Based on the phylogenetic tree, Prof. Shirai succeeded in estimating the ancestral genes of luciferase at seven branching points, dating back to 120 million years before the appearance of fireflies.
The green light of a long-awaited dream being fulfilled
Revival of a sight from ancient history
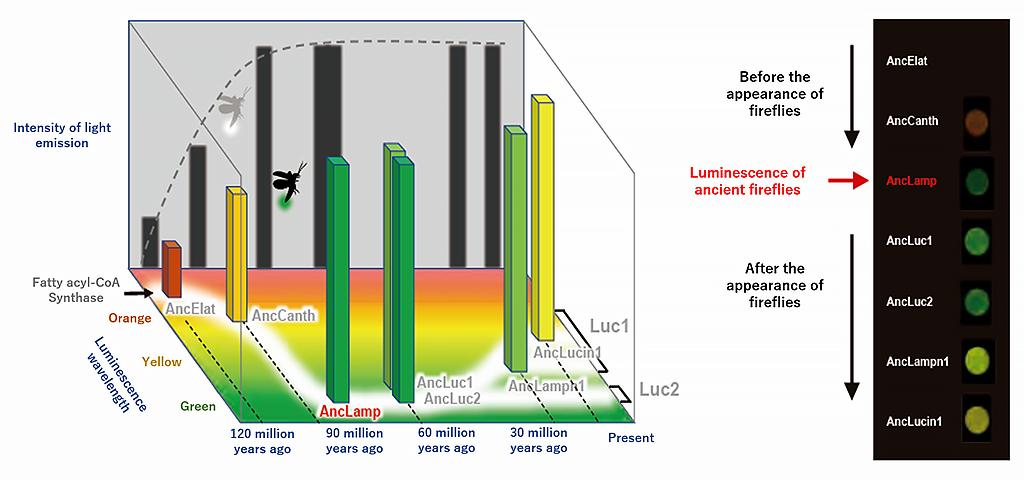
Based on the 100-million-year-old DNA sequence estimated by Prof. Shirai, Prof. Oba reproduced the ancestral DNA of luciferase by chemical synthesis. This ancestral DNA was synthesized by incorporating it into the DNA of Escherichia coli and then subsequently made to interact with luciferin. This resulted in a deep green light being emitted (Fig 4).

"I was really happy that my long-held dream had come true when I realized that the light I was looking at was emitted by the ancestors of fireflies during the Cretaceous period, when dinosaurs still walked the earth," said Prof. Oba, explaining how impressed he was at the time. Prof. Oba recalled how he was worried about whether he could get the protein to luminesce at all but confessed that his group expected that it would shine green. "Green is the most visibly distinct color for nocturnal animals. It is likely that these ancestral animals lit up distinctly in order to give a conspicuous warning to predators that they were poisonous, or as if to say, "I taste bad!" Prof. Oba explained.
Continuing from there, analysis of the luciferase genes at each of the branch points clarified the color of the luminescence of each gene, with the ancestral Fatty acyl-CoA synthetase emitting a red/yellow glow, and after the appearance of the firefly this shifted in steps from deep green, to yellow-green, yellow, and orange (Fig 5).
Information on the luciferase genes that were not used in this analysis is currently still being decoded.
Prof. Oba continued to speak about future prospects, "The more detailed this molecular evolutionary phylogeny is, the greater the accuracy of estimation becomes, so as new information is decoded it is being added in sequence. On top of the 7 branch points we revealed in this research, we estimate that there are more ancestral genes, and I would like to investigate the evolution of luciferase in even more detail." If the details of this evolution become more clear, it will likely become possible to also simulate evolutionary steps that didn't occur. Up to this point, artificial luciferases of varying color have been developed through random changes in the sequence of amino acids. However, this research has clarified the evolutionary process that leads to a change in emitted color, and there are expectations that this knowledge can be used in the targeted design of new artificial luciferases without randomness.

Prof. Shirai also felt that his joint research with Prof. Oba was highly worthwhile. "The significance and effectiveness of ancestral sequence estimation have been conveyed to many people. Maximum likelihood is a powerful tool that can be scientifically traced back in history and reproduced and verified. I would also like to elucidate the evolutionary process of other organisms," he emphasized strongly.
Until now, these creatures could only be seen in the form of fossils and the color of their luminescence was lost to history. It is no exaggeration to state that these researchers have succeeded in reviving some part of an ancient scene in color but they have only just begun to review and reveal the processes of evolution through the merging of genetic engineering and computational science.
We await their next discoveries with bated breath.

The image shows a photophore at the tail end, but of course the light emission color is not retained in the fossil.
(Imaging donation: Dr. Kazantsev Moscow Insect Center)




